Gene
KWMTBOMO02025 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002953
Annotation
PREDICTED:_probable_26S_proteasome_non-ATPase_regulatory_subunit_3_[Amyelois_transitella]
Full name
Probable 26S proteasome non-ATPase regulatory subunit 3
Alternative Name
26S proteasome regulatory subunit RPN3
Diphenol oxidase A2 component
Diphenol oxidase A2 component
Location in the cell
Nuclear Reliability : 2.306
Sequence
CDS
ATGGCTCCAGTGGAAGATGTTGAAATGAAAAATGTGGAAAGTCCTGCAGCTGTTAGTGACGTTGAGACAGCGGATGTAAAGAAAGATGCTGATGTACTAGCTGTTCAGGATCTACGAGAACACATCAGACAAATCGATAAAGCTGTTACTTCGAAAGAACCTAGATTCGCTATGCGTGTACTTAGAGCACTGCCGAGCACTCGCAGGAAGTTAAACGGTAACGTTCTGCGAGCTATTATCAATCAGATTTATCCCGCTAGTGCTGAGAAAGAAACCATACTATCTTTTGTGGAAAATCCTCTCCCTGGTGCCGTAGAAATAGAAGCTCCCAGGTCACGAAGCGCCCCTAAGCAGCCAGTGCCCGAAGTGGATACATATTTGCATCTCCTTGTGCTGTTACGTTTGTTGGATACAAACAAACTTGAAGAAGCCACAGAATGCTCTCAGCAACTAATGACAAAAGTTACTACTCAAAATCGAAGAACCCTTGACTTGATTGCTGCAAAATGCTATTTTTATCACTCAAGAGTGTTTGAACTAACTAACAAGTTGGATGTGATCAGAGGTCTCTTACATGCCCGACTTCGTACATCAACATTGAGAAATGACTATGAAGGTCAAGCTGTTATTATCAACTGCCTGTTGAGAAATTACTTGCACTACTCTTTGTATGATCAAGCCGATAAATTAGTAAGTAAGTCAGTTTTTCCTGAAAATGCCAGCAACAATGAGTGGGCAAGATTTCTGTACTACCTTGGCAGAATAAAAGCAGCAAGGTTAGAATACAGTGATGCACACAAACATCTAGTGCAGGCTTTAAGAAAAGCTCCCCAAACAGCTGCTGTTGGATTCCGTCAAACGGTCCAAAAATTAGCAATTGTAGTGGAATTGCTATTAGGTGATATACCGGAGCGAGTGATTTTCCGTCAAGCTCCACTTCGTAAAGCATTAGCTCCATATTTCCAGCTCACCCAAGCTGTGCGTCTTGGAAACTTGCAAAGATTTGGTGAAGTATTGGAGAATTATGGGCCACAGTTCCGAAATGATCACACATTTACATTGATCCTTCGTCTTAGACAGAATGTTATTAAGACTGCTATTCGTTCAATTGGATTGTCCTATTCTCGCATATCACCAAAAGATATAGCTCGCAAATTAGGGCTTGACTCTGCTGAAGATGCAGAATTCATTGTTGCTAAGGCAATCAGAGATGGAGTGATTGAGGCAACATTAGACCCTGTAAAGGGCTACATGAGTAATAAAGAAAGCTCAGATATTTATTGTACTAGAGAGCCACAGTTAGCCTTTCATCAGCGGATTTCATTTTGCCTAGATTTACACAACCAAAGTGTTAAGGCAATGAGATATCCACCAAAATCATACGGAAAAGAATTGGAAAGTGCTGAGGAACGCCGTGAAAGAGAACAGCAAGATCTGGAACTTGCCAAAGAAATGGCAGAGGAAGATGATGATGGTTTCCCATGA
Protein
MAPVEDVEMKNVESPAAVSDVETADVKKDADVLAVQDLREHIRQIDKAVTSKEPRFAMRVLRALPSTRRKLNGNVLRAIINQIYPASAEKETILSFVENPLPGAVEIEAPRSRSAPKQPVPEVDTYLHLLVLLRLLDTNKLEEATECSQQLMTKVTTQNRRTLDLIAAKCYFYHSRVFELTNKLDVIRGLLHARLRTSTLRNDYEGQAVIINCLLRNYLHYSLYDQADKLVSKSVFPENASNNEWARFLYYLGRIKAARLEYSDAHKHLVQALRKAPQTAAVGFRQTVQKLAIVVELLLGDIPERVIFRQAPLRKALAPYFQLTQAVRLGNLQRFGEVLENYGPQFRNDHTFTLILRLRQNVIKTAIRSIGLSYSRISPKDIARKLGLDSAEDAEFIVAKAIRDGVIEATLDPVKGYMSNKESSDIYCTREPQLAFHQRISFCLDLHNQSVKAMRYPPKSYGKELESAEERREREQQDLELAKEMAEEDDDGFP
Summary
Description
Acts as a regulatory subunit of the 26 proteasome which is involved in the ATP-dependent degradation of ubiquitinated proteins.
Subunit
The 26S proteasome is composed of a core protease, known as the 20S proteasome, capped at one or both ends by the 19S regulatory complex (RC). The RC is composed of at least 18 different subunits in two subcomplexes, the base and the lid, which form the portions proximal and distal to the 20S proteolytic core, respectively (By similarity).
Similarity
Belongs to the proteasome subunit S3 family.
Keywords
Complete proteome
Proteasome
Reference proteome
Feature
chain Probable 26S proteasome non-ATPase regulatory subunit 3
Uniprot
H9J0B7
A0A2A4JY67
A0A2H1X096
A0A1E1WF21
I4DK33
A0A194RD45
+ More
S4PGF7 A0A212FA38 A0A0L7LEF5 A0A2P8XTW6 V5GN64 A0A0C5AST9 A0A1B0DLH7 A0A2J7QBQ9 A0A1B6DCY0 B0WCQ3 A0A182K3Q3 A0A1Q3FAL3 A0A1B6FCJ6 N6THH0 A0A182R9S0 U4U1Q6 A0A067QNT9 A0A182MIP1 A0A182NEV7 A0A182PDF1 A0A023EU98 A0A2M4ADN7 Q16N14 A0A2M3ZH99 A0A182F5I5 W5JD00 A0A182YFG0 A0A182WGX4 A0A2M4BLH4 A0A182H859 U5EZB6 D6WL27 A0A182JBK4 A0A084VMC7 A0A182QVK3 A0A336KQ90 A0A1Y1MR71 A0A1L8DT32 A0A182VCR5 A0A182TT01 A0A182X2Y2 A0A182I4X2 O61470 A0A336K9E6 A0A232FC54 K7IPX0 A0A1J1I086 A0A0V0G3R3 A0A023FCV2 A0A069DU15 E2BQ36 A0A0P4VL30 A0A158NV98 A0A0A9Y084 A0A195B8F8 A0A0C9RE25 A0A0N0BHA9 E0VFN6 A0A195FV61 A0A026WTC0 E9IDJ2 E2AFJ6 A0A195ED72 A0A0K8WG40 A0A0J7L773 A0A034WS53 A0A2A3ET91 A0A154PIE7 A0A310SLR7 D3TN58 A0A1A9V9I3 A0A1W4XAH9 A0A1B0A540 A0A1A9W063 Q9U5Z8 A0A1B0G931 T1I033 A0A2R7W1Z7 A0A0L7RGT6 A0A088ASB8 W8C3L9 A0A1B0B7N7 A0A1A9XL20 A0A151IPB9 R4WQT4 B4N147 B4LS14 F4WNN9 A0A3B0JVU4 B4JAM3 Q29PI4 B4GKT7 A0A1I8MWP0 A0A1W4WA38
S4PGF7 A0A212FA38 A0A0L7LEF5 A0A2P8XTW6 V5GN64 A0A0C5AST9 A0A1B0DLH7 A0A2J7QBQ9 A0A1B6DCY0 B0WCQ3 A0A182K3Q3 A0A1Q3FAL3 A0A1B6FCJ6 N6THH0 A0A182R9S0 U4U1Q6 A0A067QNT9 A0A182MIP1 A0A182NEV7 A0A182PDF1 A0A023EU98 A0A2M4ADN7 Q16N14 A0A2M3ZH99 A0A182F5I5 W5JD00 A0A182YFG0 A0A182WGX4 A0A2M4BLH4 A0A182H859 U5EZB6 D6WL27 A0A182JBK4 A0A084VMC7 A0A182QVK3 A0A336KQ90 A0A1Y1MR71 A0A1L8DT32 A0A182VCR5 A0A182TT01 A0A182X2Y2 A0A182I4X2 O61470 A0A336K9E6 A0A232FC54 K7IPX0 A0A1J1I086 A0A0V0G3R3 A0A023FCV2 A0A069DU15 E2BQ36 A0A0P4VL30 A0A158NV98 A0A0A9Y084 A0A195B8F8 A0A0C9RE25 A0A0N0BHA9 E0VFN6 A0A195FV61 A0A026WTC0 E9IDJ2 E2AFJ6 A0A195ED72 A0A0K8WG40 A0A0J7L773 A0A034WS53 A0A2A3ET91 A0A154PIE7 A0A310SLR7 D3TN58 A0A1A9V9I3 A0A1W4XAH9 A0A1B0A540 A0A1A9W063 Q9U5Z8 A0A1B0G931 T1I033 A0A2R7W1Z7 A0A0L7RGT6 A0A088ASB8 W8C3L9 A0A1B0B7N7 A0A1A9XL20 A0A151IPB9 R4WQT4 B4N147 B4LS14 F4WNN9 A0A3B0JVU4 B4JAM3 Q29PI4 B4GKT7 A0A1I8MWP0 A0A1W4WA38
Pubmed
19121390
22651552
26354079
23622113
22118469
26227816
+ More
29403074 25653432 29136191 23537049 24845553 24945155 17510324 20920257 23761445 25244985 26483478 18362917 19820115 24438588 28004739 9988317 12364791 28648823 20075255 25474469 26334808 20798317 27129103 21347285 25401762 26823975 20566863 24508170 30249741 21282665 25348373 20353571 10835480 24495485 23691247 17994087 21719571 15632085 25315136
29403074 25653432 29136191 23537049 24845553 24945155 17510324 20920257 23761445 25244985 26483478 18362917 19820115 24438588 28004739 9988317 12364791 28648823 20075255 25474469 26334808 20798317 27129103 21347285 25401762 26823975 20566863 24508170 30249741 21282665 25348373 20353571 10835480 24495485 23691247 17994087 21719571 15632085 25315136
EMBL
BABH01010598
NWSH01000407
PCG76618.1
ODYU01012450
SOQ58759.1
GDQN01005527
+ More
JAT85527.1 AK401651 KQ459601 BAM18273.1 KPI93769.1 KQ460597 KPJ13831.1 GAIX01003697 JAA88863.1 AGBW02009527 OWR50594.1 JTDY01001441 KOB73852.1 PYGN01001356 PSN35451.1 GALX01005464 JAB63002.1 KM999967 QKKF02030719 AJK90582.1 RZF34685.1 AJVK01006652 NEVH01016296 PNF26024.1 GEDC01013747 JAS23551.1 DS231890 EDS43779.1 GFDL01010421 JAV24624.1 GECZ01021837 JAS47932.1 APGK01037110 KB740948 ENN77223.1 KB631899 ERL86992.1 KK853129 KDR10975.1 AXCM01001875 GAPW01001112 JAC12486.1 GGFK01005576 MBW38897.1 CH477842 EAT35739.1 GGFM01007151 MBW27902.1 ADMH02001866 ETN60770.1 GGFJ01004672 MBW53813.1 JXUM01028759 KQ560831 KXJ80696.1 GANO01001087 JAB58784.1 KQ971343 EFA04054.1 ATLV01014585 KE524975 KFB39121.1 AXCN02000840 UFQS01000634 UFQT01000634 SSX05595.1 SSX25954.1 GEZM01025134 JAV87568.1 GFDF01004537 JAV09547.1 APCN01003737 AF042732 AAAB01008984 UFQS01000222 UFQT01000222 SSX01576.1 SSX21956.1 NNAY01000436 OXU28406.1 CVRI01000037 CRK93677.1 GECL01003420 JAP02704.1 GBBI01000213 JAC18499.1 GBGD01001286 JAC87603.1 GL449694 EFN82224.1 GDKW01001493 JAI55102.1 ADTU01002880 GBHO01020659 GBHO01020658 GBRD01007483 GDHC01000011 JAG22945.1 JAG22946.1 JAG58338.1 JAQ18618.1 KQ976565 KYM80527.1 GBYB01014809 JAG84576.1 KQ435756 KOX75984.1 DS235123 EEB12192.1 KQ981215 KYN44530.1 KK107109 QOIP01000008 EZA59310.1 RLU19538.1 GL762454 EFZ21345.1 GL439118 EFN67747.1 KQ979074 KYN22772.1 GDHF01002183 JAI50131.1 LBMM01000342 LBMM01000325 KMR00160.1 KMR01328.1 GAKP01001478 JAC57474.1 KZ288191 PBC34426.1 KQ434905 KZC11264.1 KQ761155 OAD58138.1 OAD58139.1 EZ422860 ADD19136.1 AJ250874 CCAG010004816 ACPB03012597 KK854249 PTY13702.1 KQ414592 KOC70192.1 GAMC01002712 JAC03844.1 JXJN01009696 KQ976874 KYN07686.1 AK418031 BAN21246.1 CH963920 EDW78137.1 CH940649 EDW63690.1 GL888237 EGI64232.1 OUUW01000010 SPP86217.1 CH916368 EDW02809.1 CH379058 EAL34309.1 CH479184 EDW37253.1
JAT85527.1 AK401651 KQ459601 BAM18273.1 KPI93769.1 KQ460597 KPJ13831.1 GAIX01003697 JAA88863.1 AGBW02009527 OWR50594.1 JTDY01001441 KOB73852.1 PYGN01001356 PSN35451.1 GALX01005464 JAB63002.1 KM999967 QKKF02030719 AJK90582.1 RZF34685.1 AJVK01006652 NEVH01016296 PNF26024.1 GEDC01013747 JAS23551.1 DS231890 EDS43779.1 GFDL01010421 JAV24624.1 GECZ01021837 JAS47932.1 APGK01037110 KB740948 ENN77223.1 KB631899 ERL86992.1 KK853129 KDR10975.1 AXCM01001875 GAPW01001112 JAC12486.1 GGFK01005576 MBW38897.1 CH477842 EAT35739.1 GGFM01007151 MBW27902.1 ADMH02001866 ETN60770.1 GGFJ01004672 MBW53813.1 JXUM01028759 KQ560831 KXJ80696.1 GANO01001087 JAB58784.1 KQ971343 EFA04054.1 ATLV01014585 KE524975 KFB39121.1 AXCN02000840 UFQS01000634 UFQT01000634 SSX05595.1 SSX25954.1 GEZM01025134 JAV87568.1 GFDF01004537 JAV09547.1 APCN01003737 AF042732 AAAB01008984 UFQS01000222 UFQT01000222 SSX01576.1 SSX21956.1 NNAY01000436 OXU28406.1 CVRI01000037 CRK93677.1 GECL01003420 JAP02704.1 GBBI01000213 JAC18499.1 GBGD01001286 JAC87603.1 GL449694 EFN82224.1 GDKW01001493 JAI55102.1 ADTU01002880 GBHO01020659 GBHO01020658 GBRD01007483 GDHC01000011 JAG22945.1 JAG22946.1 JAG58338.1 JAQ18618.1 KQ976565 KYM80527.1 GBYB01014809 JAG84576.1 KQ435756 KOX75984.1 DS235123 EEB12192.1 KQ981215 KYN44530.1 KK107109 QOIP01000008 EZA59310.1 RLU19538.1 GL762454 EFZ21345.1 GL439118 EFN67747.1 KQ979074 KYN22772.1 GDHF01002183 JAI50131.1 LBMM01000342 LBMM01000325 KMR00160.1 KMR01328.1 GAKP01001478 JAC57474.1 KZ288191 PBC34426.1 KQ434905 KZC11264.1 KQ761155 OAD58138.1 OAD58139.1 EZ422860 ADD19136.1 AJ250874 CCAG010004816 ACPB03012597 KK854249 PTY13702.1 KQ414592 KOC70192.1 GAMC01002712 JAC03844.1 JXJN01009696 KQ976874 KYN07686.1 AK418031 BAN21246.1 CH963920 EDW78137.1 CH940649 EDW63690.1 GL888237 EGI64232.1 OUUW01000010 SPP86217.1 CH916368 EDW02809.1 CH379058 EAL34309.1 CH479184 EDW37253.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000245037 UP000291343 UP000092462 UP000235965 UP000002320 UP000075881 UP000019118 UP000075900 UP000030742 UP000027135 UP000075883 UP000075884 UP000075885 UP000008820 UP000069272 UP000000673 UP000076408 UP000075920 UP000069940 UP000249989 UP000007266 UP000075880 UP000030765 UP000075886 UP000075903 UP000075902 UP000076407 UP000075840 UP000007062 UP000215335 UP000002358 UP000183832 UP000008237 UP000005205 UP000078540 UP000053105 UP000009046 UP000078541 UP000053097 UP000279307 UP000000311 UP000078492 UP000036403 UP000242457 UP000076502 UP000078200 UP000192223 UP000092445 UP000091820 UP000092444 UP000015103 UP000053825 UP000005203 UP000092460 UP000092443 UP000078542 UP000007798 UP000008792 UP000007755 UP000268350 UP000001070 UP000001819 UP000008744 UP000095301 UP000192221
UP000245037 UP000291343 UP000092462 UP000235965 UP000002320 UP000075881 UP000019118 UP000075900 UP000030742 UP000027135 UP000075883 UP000075884 UP000075885 UP000008820 UP000069272 UP000000673 UP000076408 UP000075920 UP000069940 UP000249989 UP000007266 UP000075880 UP000030765 UP000075886 UP000075903 UP000075902 UP000076407 UP000075840 UP000007062 UP000215335 UP000002358 UP000183832 UP000008237 UP000005205 UP000078540 UP000053105 UP000009046 UP000078541 UP000053097 UP000279307 UP000000311 UP000078492 UP000036403 UP000242457 UP000076502 UP000078200 UP000192223 UP000092445 UP000091820 UP000092444 UP000015103 UP000053825 UP000005203 UP000092460 UP000092443 UP000078542 UP000007798 UP000008792 UP000007755 UP000268350 UP000001070 UP000001819 UP000008744 UP000095301 UP000192221
Interpro
SUPFAM
SSF46785
SSF46785
ProteinModelPortal
H9J0B7
A0A2A4JY67
A0A2H1X096
A0A1E1WF21
I4DK33
A0A194RD45
+ More
S4PGF7 A0A212FA38 A0A0L7LEF5 A0A2P8XTW6 V5GN64 A0A0C5AST9 A0A1B0DLH7 A0A2J7QBQ9 A0A1B6DCY0 B0WCQ3 A0A182K3Q3 A0A1Q3FAL3 A0A1B6FCJ6 N6THH0 A0A182R9S0 U4U1Q6 A0A067QNT9 A0A182MIP1 A0A182NEV7 A0A182PDF1 A0A023EU98 A0A2M4ADN7 Q16N14 A0A2M3ZH99 A0A182F5I5 W5JD00 A0A182YFG0 A0A182WGX4 A0A2M4BLH4 A0A182H859 U5EZB6 D6WL27 A0A182JBK4 A0A084VMC7 A0A182QVK3 A0A336KQ90 A0A1Y1MR71 A0A1L8DT32 A0A182VCR5 A0A182TT01 A0A182X2Y2 A0A182I4X2 O61470 A0A336K9E6 A0A232FC54 K7IPX0 A0A1J1I086 A0A0V0G3R3 A0A023FCV2 A0A069DU15 E2BQ36 A0A0P4VL30 A0A158NV98 A0A0A9Y084 A0A195B8F8 A0A0C9RE25 A0A0N0BHA9 E0VFN6 A0A195FV61 A0A026WTC0 E9IDJ2 E2AFJ6 A0A195ED72 A0A0K8WG40 A0A0J7L773 A0A034WS53 A0A2A3ET91 A0A154PIE7 A0A310SLR7 D3TN58 A0A1A9V9I3 A0A1W4XAH9 A0A1B0A540 A0A1A9W063 Q9U5Z8 A0A1B0G931 T1I033 A0A2R7W1Z7 A0A0L7RGT6 A0A088ASB8 W8C3L9 A0A1B0B7N7 A0A1A9XL20 A0A151IPB9 R4WQT4 B4N147 B4LS14 F4WNN9 A0A3B0JVU4 B4JAM3 Q29PI4 B4GKT7 A0A1I8MWP0 A0A1W4WA38
S4PGF7 A0A212FA38 A0A0L7LEF5 A0A2P8XTW6 V5GN64 A0A0C5AST9 A0A1B0DLH7 A0A2J7QBQ9 A0A1B6DCY0 B0WCQ3 A0A182K3Q3 A0A1Q3FAL3 A0A1B6FCJ6 N6THH0 A0A182R9S0 U4U1Q6 A0A067QNT9 A0A182MIP1 A0A182NEV7 A0A182PDF1 A0A023EU98 A0A2M4ADN7 Q16N14 A0A2M3ZH99 A0A182F5I5 W5JD00 A0A182YFG0 A0A182WGX4 A0A2M4BLH4 A0A182H859 U5EZB6 D6WL27 A0A182JBK4 A0A084VMC7 A0A182QVK3 A0A336KQ90 A0A1Y1MR71 A0A1L8DT32 A0A182VCR5 A0A182TT01 A0A182X2Y2 A0A182I4X2 O61470 A0A336K9E6 A0A232FC54 K7IPX0 A0A1J1I086 A0A0V0G3R3 A0A023FCV2 A0A069DU15 E2BQ36 A0A0P4VL30 A0A158NV98 A0A0A9Y084 A0A195B8F8 A0A0C9RE25 A0A0N0BHA9 E0VFN6 A0A195FV61 A0A026WTC0 E9IDJ2 E2AFJ6 A0A195ED72 A0A0K8WG40 A0A0J7L773 A0A034WS53 A0A2A3ET91 A0A154PIE7 A0A310SLR7 D3TN58 A0A1A9V9I3 A0A1W4XAH9 A0A1B0A540 A0A1A9W063 Q9U5Z8 A0A1B0G931 T1I033 A0A2R7W1Z7 A0A0L7RGT6 A0A088ASB8 W8C3L9 A0A1B0B7N7 A0A1A9XL20 A0A151IPB9 R4WQT4 B4N147 B4LS14 F4WNN9 A0A3B0JVU4 B4JAM3 Q29PI4 B4GKT7 A0A1I8MWP0 A0A1W4WA38
PDB
6EPF
E-value=7.50328e-146,
Score=1327
Ontologies
GO
PANTHER
Topology
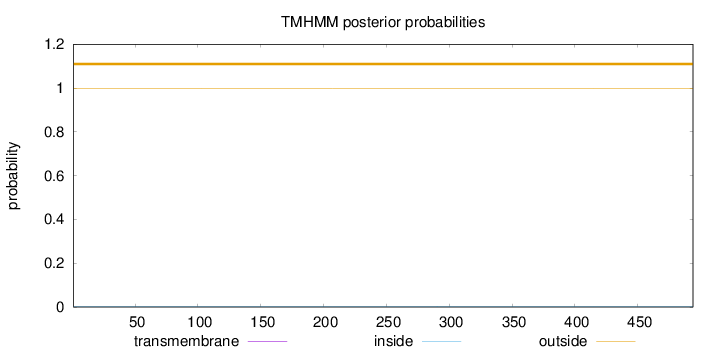
Length:
494
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000990000000000002
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00198
outside
1 - 494
Population Genetic Test Statistics
Pi
141.965908
Theta
172.770132
Tajima's D
-0.575928
CLR
666.897229
CSRT
0.223038848057597
Interpretation
Uncertain
