Pre Gene Modal
BGIBMGA013570
Annotation
Uncharacterized_protein_OBRU01_11172_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 2.169
Sequence
CDS
ATGCTTAAACGCTTTTGTGTGTTGTTTATAGCGCTTGCTGTATTTGGTGAATGTTCAGTAACAACCTTCCCAAAGACTCATACACAACCAGAATGGGAAGCCCACGTGAAAGACTTTATAAAATTGATCGACGTTCAGAAATTAATGAGTATTGCGGTTAACTACATTTCCGATCCAGAAGTTATGAACTTCGTCGAGTTCCTGTTAAGTCCGGAATTCAAGAAAATAATTTGGGAAATAGAAATCATGCCAGAGTTCAGAGAAGCTTATGGATATTTGAAAGTCAAGGGCTATGATTTTGGATTCGCGTTCGACTTTATTAATAACGCTTTAAATATGCCGCCGTTTGAACCAAAAACACCATTGAGGAATAAAAAAGGAATATCTGGAGTGTTTGAAGAAATGATGGCTGCGTTGCCCTTTGAAGAAATGAAAAAACTTTTCGAAGATAAACTAGAGACGAGTACCGAAATTTCCGAACTCTTCAAGATTATCAATTCTGAAGAATATTTGTCAATTATAAAGAGGATGTCCGCTAATCAAGAGTTTATTGAAATTAAGAATATTTTAACATTATACGGATTTAAATTTGATTATATATGTTCGTTTGCTAAGGGTATCTTCGGCGAATATTATCAGGGTATCTTTTGTGCTTAA
Protein
MLKRFCVLFIALAVFGECSVTTFPKTHTQPEWEAHVKDFIKLIDVQKLMSIAVNYISDPEVMNFVEFLLSPEFKKIIWEIEIMPEFREAYGYLKVKGYDFGFAFDFINNALNMPPFEPKTPLRNKKGISGVFEEMMAALPFEEMKKLFEDKLETSTEISELFKIINSEEYLSIIKRMSANQEFIEIKNILTLYGFKFDYICSFAKGIFGEYYQGIFCA
Summary
Uniprot
H9JVK7
A0A0L7LCH4
A0A212ERR5
A0A2P0XIZ6
O18535
Q9TZR6
+ More
O18528 O18530 D3JUE8 A0A067QVM1 A0A2P8YVU1 O96522 A9XFX0 O96523 Q9UAM5 A0A2P8Y6L1 A0A0C9PIB4 A0A2J7R8A2 A0A2J7R8A9 K7J3S2 A0A182IUC0 B0X1R6 F4MI47 A0A154NZ83 E2A6C9 A8CW22 A0A0L7R8C9 A0A0N0BIK3 A0A026WX70 A0A0L7R8K3 A0A087ZXS1 B0X1R3 F4X027 B0X1R2 B0WED5 A0A2P8XBP4 A0A2A3E602 A0A195CV20 A0A0M9A7T2 A0A088AIT4 E9J3M2 A0A2P8YX68 A0A151XFV9 E2BBE3 A0A087ZXS3 A0A087ZXS2 A0A151JU60 A0A2A3E574 A0A195E8S7 Q16K34 A0A1W7R7P8 A0A2A3E6L0 A0A158NFC6 A0A195BI59 A0A0L7R885 A0A0L7R882 A0A026WXX3 U5EMJ5 Q8T5C5 A0A1S4G7T6 Q16IQ0 A0A182Q2Q9 Q9GPE6 Q16WN9 A0A1L8DR29 Q16K33 A0A0L7R8C5 A0A084W5V9 A0A1L8DQZ4 A0A1L8DRD7 Q8WT62 Q16SZ6 A0A3L8E2R5 Q16K36 A0A3G5BIJ4 A0A0J7NFX7 Q16SZ5 A0A1L8DRE6 A0A3S2L044 Q16WN7 F4MI43 A0A1L8DR85 A0A182VSV6 Q9GPE7 A0A1S4FS07 A0A182G0S0 A0A1B0GLP4 A0A182G6A6 T1EAR6 Q16RL1 A0A023ELN6 Q17KR2 A8C9U5 A0A182IU77 E9J3L7 A8CW39 A0A1B0EWH2
O18528 O18530 D3JUE8 A0A067QVM1 A0A2P8YVU1 O96522 A9XFX0 O96523 Q9UAM5 A0A2P8Y6L1 A0A0C9PIB4 A0A2J7R8A2 A0A2J7R8A9 K7J3S2 A0A182IUC0 B0X1R6 F4MI47 A0A154NZ83 E2A6C9 A8CW22 A0A0L7R8C9 A0A0N0BIK3 A0A026WX70 A0A0L7R8K3 A0A087ZXS1 B0X1R3 F4X027 B0X1R2 B0WED5 A0A2P8XBP4 A0A2A3E602 A0A195CV20 A0A0M9A7T2 A0A088AIT4 E9J3M2 A0A2P8YX68 A0A151XFV9 E2BBE3 A0A087ZXS3 A0A087ZXS2 A0A151JU60 A0A2A3E574 A0A195E8S7 Q16K34 A0A1W7R7P8 A0A2A3E6L0 A0A158NFC6 A0A195BI59 A0A0L7R885 A0A0L7R882 A0A026WXX3 U5EMJ5 Q8T5C5 A0A1S4G7T6 Q16IQ0 A0A182Q2Q9 Q9GPE6 Q16WN9 A0A1L8DR29 Q16K33 A0A0L7R8C5 A0A084W5V9 A0A1L8DQZ4 A0A1L8DRD7 Q8WT62 Q16SZ6 A0A3L8E2R5 Q16K36 A0A3G5BIJ4 A0A0J7NFX7 Q16SZ5 A0A1L8DRE6 A0A3S2L044 Q16WN7 F4MI43 A0A1L8DR85 A0A182VSV6 Q9GPE7 A0A1S4FS07 A0A182G0S0 A0A1B0GLP4 A0A182G6A6 T1EAR6 Q16RL1 A0A023ELN6 Q17KR2 A8C9U5 A0A182IU77 E9J3L7 A8CW39 A0A1B0EWH2
Pubmed
EMBL
BABH01039015
JTDY01001668
KOB73202.1
AGBW02012969
OWR44144.1
KY661332
+ More
AVA17419.1 U78970 AAC34312.1 AF072222 AAD13533.1 U69261 AAC34737.1 U69957 AAB82404.1 GU301883 ADB92492.1 KK853323 KDR08524.1 PYGN01000332 PSN48368.1 AF072220 AAD13531.1 EF202179 ABP04045.1 AF072221 AAD13532.1 AF072219 AAD13530.2 PYGN01000874 PSN39804.1 GBYB01000663 JAG70430.1 NEVH01006723 PNF37061.1 PNF37062.1 DS232267 EDS38794.1 EZ933295 ADJ57676.1 KQ434783 KZC04901.1 GL437123 EFN71014.1 AJWK01028468 EU124571 ABV60289.1 KQ414633 KOC67099.1 KQ435729 KOX77672.1 KK107077 QOIP01000001 EZA60607.1 RLU26326.1 RLU26700.1 KOC67096.1 EDS38791.1 GL888486 EGI60214.1 EDS38790.1 DS231908 EDS45562.1 PYGN01003836 PSN29414.1 KZ288364 PBC26914.1 KQ977279 KYN04372.1 KOX77673.1 GL768066 EFZ12622.1 PYGN01000306 PSN48846.1 KQ982182 KYQ59195.1 GL447038 EFN86980.1 KQ981791 KYN35782.1 PBC26913.1 KQ979440 KYN21516.1 CH477978 CH477559 EAT34668.1 EAT38996.1 GEHC01000476 JAV47169.1 PBC26912.1 ADTU01014245 KQ976464 KYM84538.1 KOC67090.1 KOC67092.1 EZA60591.1 GANO01000999 JAB58872.1 AY050565 CH478060 CH477662 AAL05408.1 EAT34153.1 EAT34155.1 EAT37592.1 EAT34154.1 AXCN02001866 AY009156 AAG35193.1 EAT38997.1 GFDF01005162 JAV08922.1 EAT34669.1 KOC67094.1 ATLV01020699 KE525305 KFB45603.1 GFDF01005188 JAV08896.1 GFDF01005189 JAV08895.1 AY038041 AAK72505.2 EAT37594.1 RLU27030.1 EAT34666.1 MK075200 AYV99603.1 LBMM01005495 KMQ91470.1 EAT37595.1 GFDF01005164 JAV08920.1 RSAL01000319 RVE42568.1 EAT38998.1 EZ933291 ADJ57672.1 GFDF01005163 JAV08921.1 AY009155 AAG35192.1 JXUM01006083 KQ560208 KXJ83828.1 JXUM01044727 KQ561411 KXJ78663.1 GAMD01000177 JAB01414.1 CH477705 EAT37044.1 GAPW01004424 JAC09174.1 CH477222 EAT47242.1 EU031911 ABV44704.1 EFZ12575.1 EU124577 ABV60295.1
AVA17419.1 U78970 AAC34312.1 AF072222 AAD13533.1 U69261 AAC34737.1 U69957 AAB82404.1 GU301883 ADB92492.1 KK853323 KDR08524.1 PYGN01000332 PSN48368.1 AF072220 AAD13531.1 EF202179 ABP04045.1 AF072221 AAD13532.1 AF072219 AAD13530.2 PYGN01000874 PSN39804.1 GBYB01000663 JAG70430.1 NEVH01006723 PNF37061.1 PNF37062.1 DS232267 EDS38794.1 EZ933295 ADJ57676.1 KQ434783 KZC04901.1 GL437123 EFN71014.1 AJWK01028468 EU124571 ABV60289.1 KQ414633 KOC67099.1 KQ435729 KOX77672.1 KK107077 QOIP01000001 EZA60607.1 RLU26326.1 RLU26700.1 KOC67096.1 EDS38791.1 GL888486 EGI60214.1 EDS38790.1 DS231908 EDS45562.1 PYGN01003836 PSN29414.1 KZ288364 PBC26914.1 KQ977279 KYN04372.1 KOX77673.1 GL768066 EFZ12622.1 PYGN01000306 PSN48846.1 KQ982182 KYQ59195.1 GL447038 EFN86980.1 KQ981791 KYN35782.1 PBC26913.1 KQ979440 KYN21516.1 CH477978 CH477559 EAT34668.1 EAT38996.1 GEHC01000476 JAV47169.1 PBC26912.1 ADTU01014245 KQ976464 KYM84538.1 KOC67090.1 KOC67092.1 EZA60591.1 GANO01000999 JAB58872.1 AY050565 CH478060 CH477662 AAL05408.1 EAT34153.1 EAT34155.1 EAT37592.1 EAT34154.1 AXCN02001866 AY009156 AAG35193.1 EAT38997.1 GFDF01005162 JAV08922.1 EAT34669.1 KOC67094.1 ATLV01020699 KE525305 KFB45603.1 GFDF01005188 JAV08896.1 GFDF01005189 JAV08895.1 AY038041 AAK72505.2 EAT37594.1 RLU27030.1 EAT34666.1 MK075200 AYV99603.1 LBMM01005495 KMQ91470.1 EAT37595.1 GFDF01005164 JAV08920.1 RSAL01000319 RVE42568.1 EAT38998.1 EZ933291 ADJ57672.1 GFDF01005163 JAV08921.1 AY009155 AAG35192.1 JXUM01006083 KQ560208 KXJ83828.1 JXUM01044727 KQ561411 KXJ78663.1 GAMD01000177 JAB01414.1 CH477705 EAT37044.1 GAPW01004424 JAC09174.1 CH477222 EAT47242.1 EU031911 ABV44704.1 EFZ12575.1 EU124577 ABV60295.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000027135
UP000245037
UP000235965
+ More
UP000002358 UP000075880 UP000002320 UP000076502 UP000000311 UP000092461 UP000053825 UP000053105 UP000053097 UP000279307 UP000005203 UP000007755 UP000242457 UP000078542 UP000075809 UP000008237 UP000078541 UP000078492 UP000008820 UP000005205 UP000078540 UP000075886 UP000030765 UP000036403 UP000283053 UP000075920 UP000069940 UP000249989
UP000002358 UP000075880 UP000002320 UP000076502 UP000000311 UP000092461 UP000053825 UP000053105 UP000053097 UP000279307 UP000005203 UP000007755 UP000242457 UP000078542 UP000075809 UP000008237 UP000078541 UP000078492 UP000008820 UP000005205 UP000078540 UP000075886 UP000030765 UP000036403 UP000283053 UP000075920 UP000069940 UP000249989
PRIDE
Pfam
PF06757 Ins_allergen_rp
Interpro
IPR010629
Ins_allergen
ProteinModelPortal
H9JVK7
A0A0L7LCH4
A0A212ERR5
A0A2P0XIZ6
O18535
Q9TZR6
+ More
O18528 O18530 D3JUE8 A0A067QVM1 A0A2P8YVU1 O96522 A9XFX0 O96523 Q9UAM5 A0A2P8Y6L1 A0A0C9PIB4 A0A2J7R8A2 A0A2J7R8A9 K7J3S2 A0A182IUC0 B0X1R6 F4MI47 A0A154NZ83 E2A6C9 A8CW22 A0A0L7R8C9 A0A0N0BIK3 A0A026WX70 A0A0L7R8K3 A0A087ZXS1 B0X1R3 F4X027 B0X1R2 B0WED5 A0A2P8XBP4 A0A2A3E602 A0A195CV20 A0A0M9A7T2 A0A088AIT4 E9J3M2 A0A2P8YX68 A0A151XFV9 E2BBE3 A0A087ZXS3 A0A087ZXS2 A0A151JU60 A0A2A3E574 A0A195E8S7 Q16K34 A0A1W7R7P8 A0A2A3E6L0 A0A158NFC6 A0A195BI59 A0A0L7R885 A0A0L7R882 A0A026WXX3 U5EMJ5 Q8T5C5 A0A1S4G7T6 Q16IQ0 A0A182Q2Q9 Q9GPE6 Q16WN9 A0A1L8DR29 Q16K33 A0A0L7R8C5 A0A084W5V9 A0A1L8DQZ4 A0A1L8DRD7 Q8WT62 Q16SZ6 A0A3L8E2R5 Q16K36 A0A3G5BIJ4 A0A0J7NFX7 Q16SZ5 A0A1L8DRE6 A0A3S2L044 Q16WN7 F4MI43 A0A1L8DR85 A0A182VSV6 Q9GPE7 A0A1S4FS07 A0A182G0S0 A0A1B0GLP4 A0A182G6A6 T1EAR6 Q16RL1 A0A023ELN6 Q17KR2 A8C9U5 A0A182IU77 E9J3L7 A8CW39 A0A1B0EWH2
O18528 O18530 D3JUE8 A0A067QVM1 A0A2P8YVU1 O96522 A9XFX0 O96523 Q9UAM5 A0A2P8Y6L1 A0A0C9PIB4 A0A2J7R8A2 A0A2J7R8A9 K7J3S2 A0A182IUC0 B0X1R6 F4MI47 A0A154NZ83 E2A6C9 A8CW22 A0A0L7R8C9 A0A0N0BIK3 A0A026WX70 A0A0L7R8K3 A0A087ZXS1 B0X1R3 F4X027 B0X1R2 B0WED5 A0A2P8XBP4 A0A2A3E602 A0A195CV20 A0A0M9A7T2 A0A088AIT4 E9J3M2 A0A2P8YX68 A0A151XFV9 E2BBE3 A0A087ZXS3 A0A087ZXS2 A0A151JU60 A0A2A3E574 A0A195E8S7 Q16K34 A0A1W7R7P8 A0A2A3E6L0 A0A158NFC6 A0A195BI59 A0A0L7R885 A0A0L7R882 A0A026WXX3 U5EMJ5 Q8T5C5 A0A1S4G7T6 Q16IQ0 A0A182Q2Q9 Q9GPE6 Q16WN9 A0A1L8DR29 Q16K33 A0A0L7R8C5 A0A084W5V9 A0A1L8DQZ4 A0A1L8DRD7 Q8WT62 Q16SZ6 A0A3L8E2R5 Q16K36 A0A3G5BIJ4 A0A0J7NFX7 Q16SZ5 A0A1L8DRE6 A0A3S2L044 Q16WN7 F4MI43 A0A1L8DR85 A0A182VSV6 Q9GPE7 A0A1S4FS07 A0A182G0S0 A0A1B0GLP4 A0A182G6A6 T1EAR6 Q16RL1 A0A023ELN6 Q17KR2 A8C9U5 A0A182IU77 E9J3L7 A8CW39 A0A1B0EWH2
PDB
4JRB
E-value=1.17888e-19,
Score=234
Ontologies
GO
PANTHER
Topology
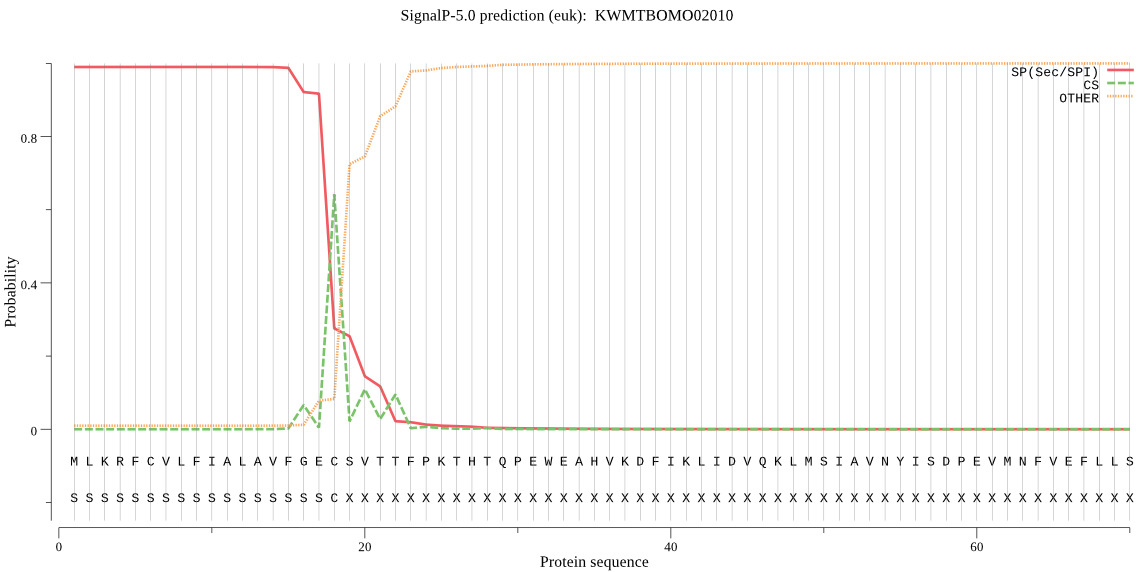
SignalP
Position: 1 - 18,
Likelihood: 0.989705
Length:
218
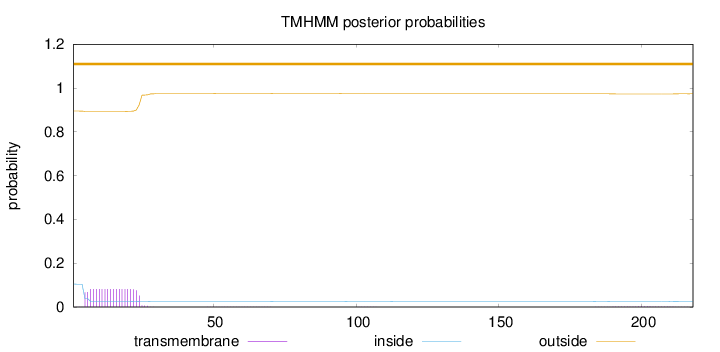
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.66043
Exp number, first 60 AAs:
1.60338
Total prob of N-in:
0.10509
outside
1 - 218
Population Genetic Test Statistics
Pi
78.128955
Theta
85.400068
Tajima's D
-0.221115
CLR
4.902734
CSRT
0.314084295785211
Interpretation
Uncertain
