Gene
KWMTBOMO02009
Pre Gene Modal
BGIBMGA013568
Annotation
osiris_21_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.264 Mitochondrial Reliability : 1.034
Sequence
CDS
ATGGATGCCGAGGATTACTCAGAGAACGAAATAGATAGTGGCAGAGCTGGGGAGAGCTCCTGGTGGCCGTCAGCCGAATTTGCCGTCTTACAGAAAGTTTATGATGACTGTAGCTCTCAGAAACATATGTCACTGTGTCTCAAGGGCAAAGCTTTGATTGCATTATCACGAGCGGTGGAGCAGGAGAGCATTCAGATCACTGATGGGTTATCGTTAGTGAAGGAGGGCGACGCTCCCTCAGCAGTAGCAGAGCCACGTTTCTTTCCAGGAATGACGAAAGAGGAGAAAATCGACTCTATGCTGCGCACTAAATTCGAGCAGTTAATGAACACTCATACAGTTTCTCTTGATCTTGCTACGGAAGGCAGAGGTAGAGGTAAAAAGGTCCTGCCTTATTTGCTCCTCGGTATCTTTTCTACCATGAGCATATTCGGAGGCATGGCATTGAAGACCCTAGCCGCTATCGCAGGCAAAGCCCTTATCGCCAGTAAAGTGGCTTTGACAATTGCAGGCATTATTGCCCTCAAGAAATTGTTCAGTCATGAGACGCCCACTGAAACGACGTTCCAAGTTCACGCAGACGGACACCATAGGCGGAATCTGTATGTGGTGAGACCAGTGAGCCCAAGTGCGGTGGACCCTTATCATTATGCTAAACCGACACTCGCTAAAGCTTAG
Protein
MDAEDYSENEIDSGRAGESSWWPSAEFAVLQKVYDDCSSQKHMSLCLKGKALIALSRAVEQESIQITDGLSLVKEGDAPSAVAEPRFFPGMTKEEKIDSMLRTKFEQLMNTHTVSLDLATEGRGRGKKVLPYLLLGIFSTMSIFGGMALKTLAAIAGKALIASKVALTIAGIIALKKLFSHETPTETTFQVHADGHHRRNLYVVRPVSPSAVDPYHYAKPTLAKA
Summary
Uniprot
B6DXA6
A0A0L7LV27
A0A437AWM1
A0A2H1WYL9
A0A194RGB9
A0A194PK17
+ More
A0A212ERM2 A0A2A4K0Q0 A0A336MM74 Q16F75 A0A1S4G3G8 W5J630 A0A182KPT6 A0A182FE35 A0A182KF03 A0A182MUB0 A0A182WMD5 A0A182QAK8 A0A182Y2W9 A0A182RUZ7 A0A182V131 A0A182UKK2 A0A182X6V8 Q7Q6E4 A0A182I225 A0A182J263 A0A182NMG6 A0A1J1IM60 A0A182PPA0 Q9VKH5 A0A1B0DLH1 A0A182F461 B4P1G1 B0WD07 B3N4I9 B0XB35 B4HX39 B4KH94 B4QAD5 A0A084WC83 A0A182GT52 B0WHK1 A0A0M3QTQ3 Q29M25 B4G6M9 W5JWV9 B4LQF7 A0A182JRE8 A0A3B0K7K7 Q17BW2 A0A0K8V355 A0A182NQ13 A0A182R913 A0A182YE31 A0A1I8M6C5 A0A182LWF4 A0A182W874 A0A182T8A1 A0A034WT89 A0A182UUV6 A0A182TJ06 A0A182XA94 A0NCW8 A0A182HX14 A0A1W4VZH9 B4JPJ0 A0A182PBU1 A0A182S8I8 A0A182J7C4 A0A182QU51 B4N7A8 A0A182LN31 B3MMU8 A0A1I8NTP6 A0A1A9ZIQ4 A0A0L0CAZ6 A0A1B0FQD7 A0A1A9Y0B8 A0A1A9WZ54 A0A1B0B0G8 A0A1A9VFJ1 W8BY59 A0A0K8UL42 A0A0A1XFD6 A0A1W4V6G8 T1GTA7 E0VVM8 A0A2R7VU74 A0A182GBT1 A0A1J1HGH6 A0A182N5P5 A0A2P8YY84 B0WLV5 A0A084VL98 Q17DB0 A0A182QTI9 A0A1S4F7C9 A0A182YGJ4
A0A212ERM2 A0A2A4K0Q0 A0A336MM74 Q16F75 A0A1S4G3G8 W5J630 A0A182KPT6 A0A182FE35 A0A182KF03 A0A182MUB0 A0A182WMD5 A0A182QAK8 A0A182Y2W9 A0A182RUZ7 A0A182V131 A0A182UKK2 A0A182X6V8 Q7Q6E4 A0A182I225 A0A182J263 A0A182NMG6 A0A1J1IM60 A0A182PPA0 Q9VKH5 A0A1B0DLH1 A0A182F461 B4P1G1 B0WD07 B3N4I9 B0XB35 B4HX39 B4KH94 B4QAD5 A0A084WC83 A0A182GT52 B0WHK1 A0A0M3QTQ3 Q29M25 B4G6M9 W5JWV9 B4LQF7 A0A182JRE8 A0A3B0K7K7 Q17BW2 A0A0K8V355 A0A182NQ13 A0A182R913 A0A182YE31 A0A1I8M6C5 A0A182LWF4 A0A182W874 A0A182T8A1 A0A034WT89 A0A182UUV6 A0A182TJ06 A0A182XA94 A0NCW8 A0A182HX14 A0A1W4VZH9 B4JPJ0 A0A182PBU1 A0A182S8I8 A0A182J7C4 A0A182QU51 B4N7A8 A0A182LN31 B3MMU8 A0A1I8NTP6 A0A1A9ZIQ4 A0A0L0CAZ6 A0A1B0FQD7 A0A1A9Y0B8 A0A1A9WZ54 A0A1B0B0G8 A0A1A9VFJ1 W8BY59 A0A0K8UL42 A0A0A1XFD6 A0A1W4V6G8 T1GTA7 E0VVM8 A0A2R7VU74 A0A182GBT1 A0A1J1HGH6 A0A182N5P5 A0A2P8YY84 B0WLV5 A0A084VL98 Q17DB0 A0A182QTI9 A0A1S4F7C9 A0A182YGJ4
Pubmed
EMBL
FJ176294
ACI23616.1
JTDY01000032
KOB79290.1
RSAL01000319
RVE42567.1
+ More
ODYU01012043 SOQ58138.1 KQ460207 KPJ16632.1 KQ459601 KPI93781.1 AGBW02012969 OWR44143.1 NWSH01000331 PCG77343.1 UFQT01000773 SSX27118.1 CH478500 EAT32888.1 ADMH02002140 ETN58320.1 AXCM01002782 AXCN02000924 AAAB01008960 EAA11788.4 APCN01000106 CVRI01000055 CRL01248.1 AE014134 AAF53094.1 AJVK01016434 CM000157 EDW88068.1 DS231892 EDS44082.1 CH954177 EDV58901.1 DS232610 EDS44005.1 CH480818 EDW52584.1 CH933807 EDW13311.1 CM000361 CM002910 EDX04690.1 KMY89750.1 ATLV01022594 KE525334 KFB47827.1 JXUM01017677 KQ560490 KXJ82139.1 DS231937 EDS27815.1 CP012523 ALC39313.1 CH379060 EAL33870.1 CH479180 EDW29143.1 ADMH02000103 ETN67815.1 CH940649 EDW63407.1 OUUW01000004 SPP79508.1 CH477315 EAT43791.1 GDHF01019284 JAI33030.1 AXCM01000100 GAKP01000216 JAC58736.1 AAAB01008849 EAU77178.2 APCN01005115 CH916372 EDV98820.1 AXCN02002279 CH964182 EDW80249.1 CH902620 EDV30973.1 JRES01000668 KNC29416.1 CCAG010013204 JXJN01006757 GAMC01000215 JAC06341.1 GDHF01025048 JAI27266.1 GBXI01004642 JAD09650.1 CAQQ02019021 DS235812 EEB17434.1 KK854089 PTY11054.1 JXUM01054123 KQ561806 KXJ77475.1 CVRI01000003 CRK87120.1 PYGN01000289 PSN49205.1 DS231991 EDS30690.1 ATLV01014432 KE524972 KFB38742.1 CH477298 EAT44319.1 AXCN02001159
ODYU01012043 SOQ58138.1 KQ460207 KPJ16632.1 KQ459601 KPI93781.1 AGBW02012969 OWR44143.1 NWSH01000331 PCG77343.1 UFQT01000773 SSX27118.1 CH478500 EAT32888.1 ADMH02002140 ETN58320.1 AXCM01002782 AXCN02000924 AAAB01008960 EAA11788.4 APCN01000106 CVRI01000055 CRL01248.1 AE014134 AAF53094.1 AJVK01016434 CM000157 EDW88068.1 DS231892 EDS44082.1 CH954177 EDV58901.1 DS232610 EDS44005.1 CH480818 EDW52584.1 CH933807 EDW13311.1 CM000361 CM002910 EDX04690.1 KMY89750.1 ATLV01022594 KE525334 KFB47827.1 JXUM01017677 KQ560490 KXJ82139.1 DS231937 EDS27815.1 CP012523 ALC39313.1 CH379060 EAL33870.1 CH479180 EDW29143.1 ADMH02000103 ETN67815.1 CH940649 EDW63407.1 OUUW01000004 SPP79508.1 CH477315 EAT43791.1 GDHF01019284 JAI33030.1 AXCM01000100 GAKP01000216 JAC58736.1 AAAB01008849 EAU77178.2 APCN01005115 CH916372 EDV98820.1 AXCN02002279 CH964182 EDW80249.1 CH902620 EDV30973.1 JRES01000668 KNC29416.1 CCAG010013204 JXJN01006757 GAMC01000215 JAC06341.1 GDHF01025048 JAI27266.1 GBXI01004642 JAD09650.1 CAQQ02019021 DS235812 EEB17434.1 KK854089 PTY11054.1 JXUM01054123 KQ561806 KXJ77475.1 CVRI01000003 CRK87120.1 PYGN01000289 PSN49205.1 DS231991 EDS30690.1 ATLV01014432 KE524972 KFB38742.1 CH477298 EAT44319.1 AXCN02001159
Proteomes
UP000037510
UP000283053
UP000053240
UP000053268
UP000007151
UP000218220
+ More
UP000008820 UP000000673 UP000075882 UP000069272 UP000075881 UP000075883 UP000075920 UP000075886 UP000076408 UP000075900 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000075880 UP000075884 UP000183832 UP000075885 UP000000803 UP000092462 UP000002282 UP000002320 UP000008711 UP000001292 UP000009192 UP000000304 UP000030765 UP000069940 UP000249989 UP000092553 UP000001819 UP000008744 UP000008792 UP000268350 UP000095301 UP000075901 UP000192221 UP000001070 UP000007798 UP000007801 UP000095300 UP000092445 UP000037069 UP000092444 UP000092443 UP000091820 UP000092460 UP000078200 UP000015102 UP000009046 UP000245037
UP000008820 UP000000673 UP000075882 UP000069272 UP000075881 UP000075883 UP000075920 UP000075886 UP000076408 UP000075900 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000075880 UP000075884 UP000183832 UP000075885 UP000000803 UP000092462 UP000002282 UP000002320 UP000008711 UP000001292 UP000009192 UP000000304 UP000030765 UP000069940 UP000249989 UP000092553 UP000001819 UP000008744 UP000008792 UP000268350 UP000095301 UP000075901 UP000192221 UP000001070 UP000007798 UP000007801 UP000095300 UP000092445 UP000037069 UP000092444 UP000092443 UP000091820 UP000092460 UP000078200 UP000015102 UP000009046 UP000245037
Pfam
PF07898 DUF1676
Interpro
IPR012464
DUF1676
ProteinModelPortal
B6DXA6
A0A0L7LV27
A0A437AWM1
A0A2H1WYL9
A0A194RGB9
A0A194PK17
+ More
A0A212ERM2 A0A2A4K0Q0 A0A336MM74 Q16F75 A0A1S4G3G8 W5J630 A0A182KPT6 A0A182FE35 A0A182KF03 A0A182MUB0 A0A182WMD5 A0A182QAK8 A0A182Y2W9 A0A182RUZ7 A0A182V131 A0A182UKK2 A0A182X6V8 Q7Q6E4 A0A182I225 A0A182J263 A0A182NMG6 A0A1J1IM60 A0A182PPA0 Q9VKH5 A0A1B0DLH1 A0A182F461 B4P1G1 B0WD07 B3N4I9 B0XB35 B4HX39 B4KH94 B4QAD5 A0A084WC83 A0A182GT52 B0WHK1 A0A0M3QTQ3 Q29M25 B4G6M9 W5JWV9 B4LQF7 A0A182JRE8 A0A3B0K7K7 Q17BW2 A0A0K8V355 A0A182NQ13 A0A182R913 A0A182YE31 A0A1I8M6C5 A0A182LWF4 A0A182W874 A0A182T8A1 A0A034WT89 A0A182UUV6 A0A182TJ06 A0A182XA94 A0NCW8 A0A182HX14 A0A1W4VZH9 B4JPJ0 A0A182PBU1 A0A182S8I8 A0A182J7C4 A0A182QU51 B4N7A8 A0A182LN31 B3MMU8 A0A1I8NTP6 A0A1A9ZIQ4 A0A0L0CAZ6 A0A1B0FQD7 A0A1A9Y0B8 A0A1A9WZ54 A0A1B0B0G8 A0A1A9VFJ1 W8BY59 A0A0K8UL42 A0A0A1XFD6 A0A1W4V6G8 T1GTA7 E0VVM8 A0A2R7VU74 A0A182GBT1 A0A1J1HGH6 A0A182N5P5 A0A2P8YY84 B0WLV5 A0A084VL98 Q17DB0 A0A182QTI9 A0A1S4F7C9 A0A182YGJ4
A0A212ERM2 A0A2A4K0Q0 A0A336MM74 Q16F75 A0A1S4G3G8 W5J630 A0A182KPT6 A0A182FE35 A0A182KF03 A0A182MUB0 A0A182WMD5 A0A182QAK8 A0A182Y2W9 A0A182RUZ7 A0A182V131 A0A182UKK2 A0A182X6V8 Q7Q6E4 A0A182I225 A0A182J263 A0A182NMG6 A0A1J1IM60 A0A182PPA0 Q9VKH5 A0A1B0DLH1 A0A182F461 B4P1G1 B0WD07 B3N4I9 B0XB35 B4HX39 B4KH94 B4QAD5 A0A084WC83 A0A182GT52 B0WHK1 A0A0M3QTQ3 Q29M25 B4G6M9 W5JWV9 B4LQF7 A0A182JRE8 A0A3B0K7K7 Q17BW2 A0A0K8V355 A0A182NQ13 A0A182R913 A0A182YE31 A0A1I8M6C5 A0A182LWF4 A0A182W874 A0A182T8A1 A0A034WT89 A0A182UUV6 A0A182TJ06 A0A182XA94 A0NCW8 A0A182HX14 A0A1W4VZH9 B4JPJ0 A0A182PBU1 A0A182S8I8 A0A182J7C4 A0A182QU51 B4N7A8 A0A182LN31 B3MMU8 A0A1I8NTP6 A0A1A9ZIQ4 A0A0L0CAZ6 A0A1B0FQD7 A0A1A9Y0B8 A0A1A9WZ54 A0A1B0B0G8 A0A1A9VFJ1 W8BY59 A0A0K8UL42 A0A0A1XFD6 A0A1W4V6G8 T1GTA7 E0VVM8 A0A2R7VU74 A0A182GBT1 A0A1J1HGH6 A0A182N5P5 A0A2P8YY84 B0WLV5 A0A084VL98 Q17DB0 A0A182QTI9 A0A1S4F7C9 A0A182YGJ4
Ontologies
PANTHER
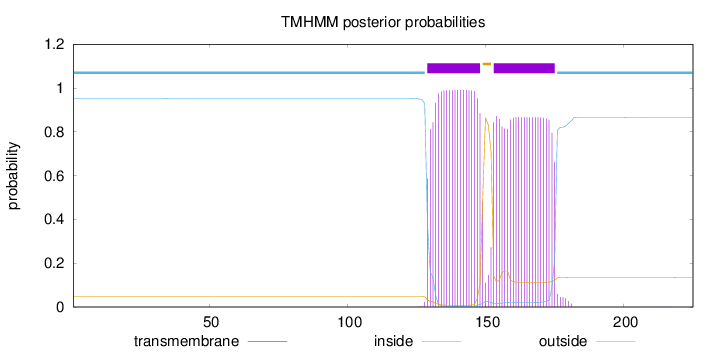
Topology
Length:
225
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
39.57591
Exp number, first 60 AAs:
0.00053
Total prob of N-in:
0.95188
inside
1 - 128
TMhelix
129 - 148
outside
149 - 152
TMhelix
153 - 175
inside
176 - 225
Population Genetic Test Statistics
Pi
288.970063
Theta
214.600115
Tajima's D
1.064086
CLR
0.067135
CSRT
0.674916254187291
Interpretation
Uncertain
