Gene
KWMTBOMO02000
Pre Gene Modal
BGIBMGA013556
Annotation
PREDICTED:_multiple_epidermal_growth_factor-like_domains_protein_8_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.831 Nuclear Reliability : 1.557
Sequence
CDS
ATGCCTAACTTCGTGGGAGATCCGGTGAACAATGTCCAGTGCGTGCCGTGCTCCTCTTATTGTTACGGACATACGCTGAAGTGCACGAACGAGGGGGAACCTGTCTCGGCGGCTGCGGAGGGTGAAGAATTCTACAGAGACGCGGCGCCGTCCACACGTGCGCGGTGCGTGCGCTGCGCTAACAACACCGACGGCCAGCGCTGCGAGCGATGTATTGAAGGATACTTCAGGGGTTCCGAGGACTTCCGAGACCCTTGCAGGCCTTGCGAATGCCACGGTCACGGCGACACGTGCGATCCCGTCACCGGCGAGAAGTGTAACTGTGGTAATAACACTGAAAGTGACGTCTCCTGTCAAACTAGCAGTTCAAGCAAGAATTCGGCACAGGAATGCTGGCGTTACCAGTGCGTGAAATGTAAGGACCTATACATGGGCGACCCGAGGAACGGACATCAATGTTATAAAACGATTAATATCGAGAATCGACTGTGCTTTGACGGGAAATCCATTGACGAATGTAAAATAAAGCCTCGACCTCTGTTCCCCGGTCAGACCGTATTCTTTGCAGTTAATCCACGTTATATGAACGTTGACATAAGAGTTATAGTGGACGTGACCCAGGGCTCTGTCGACTTATACATGAGTCCGAACGACAGTTCGTTCGTGGTCTCGGTGAACTCGAGCACAGGGGCTCACAACGTCGAGCTAGATCCGACGTACTACAAGCACGAGCCTTTCAGGAAGATGCCGACGTTCGACGAAGACGAACCCGAGAAGCCGCACATGTTCTGGTACTACGACAAGATCGAGTATCTGCTCGCGGACTACACGGCGAAGGACCTGGCAACGTACGTCGCCATAGACCGGAGGAACGTTCTGCTGCGAGTCAGGAACCTCCGCAATCGGCTCGTGCTCACCCTGCCTCATAACATACACGACCTCGCGCACACTAAGTTCTACATAGTGGTCCGTTCGCGGCACACAGACGACGGTTCCGCTAGCTACGGCGTCGTATATTTCCGTCAGGACCAACTACACATAGATCTGTTTGTGTTCTTTTCTGTGTTTTTCTCGTGCTTCTTCCTTTTCCTCGCCGCTTGTGTTGTCGCGTGGAAGGCTAAGCAGGCGGCCGACGTGCGACGCGCCCGACGAAGGCACGTGGTGGAGATGCTACATATGGCTAAACGTCCGTTCGCGGTCGTGACGGTAGTCTTAAACGGCGGAGTGACGTCATCTGGGGGACGTCGGCGCCGCGGCCCCGACCTGCGTCCTATCGCCATCGAACCCACTGGCGACGGGCGGGCCGCCGTCACTTCTGTACTCGTCAGGTTGCCCGGAGGCGCCCGCGCGCCCGTGCGGATGGCGCTAGCGTCGTCGCTGGTGGTGGTGGGGCGCACCGCCGCCCCGCGCCCCCGTCCGCGCCGCCCTCCCCCCCTCGCGCGTCTGGCCGCGCACACCTAG
Protein
MPNFVGDPVNNVQCVPCSSYCYGHTLKCTNEGEPVSAAAEGEEFYRDAAPSTRARCVRCANNTDGQRCERCIEGYFRGSEDFRDPCRPCECHGHGDTCDPVTGEKCNCGNNTESDVSCQTSSSSKNSAQECWRYQCVKCKDLYMGDPRNGHQCYKTINIENRLCFDGKSIDECKIKPRPLFPGQTVFFAVNPRYMNVDIRVIVDVTQGSVDLYMSPNDSSFVVSVNSSTGAHNVELDPTYYKHEPFRKMPTFDEDEPEKPHMFWYYDKIEYLLADYTAKDLATYVAIDRRNVLLRVRNLRNRLVLTLPHNIHDLAHTKFYIVVRSRHTDDGSASYGVVYFRQDQLHIDLFVFFSVFFSCFFLFLAACVVAWKAKQAADVRRARRRHVVEMLHMAKRPFAVVTVVLNGGVTSSGGRRRRGPDLRPIAIEPTGDGRAAVTSVLVRLPGGARAPVRMALASSLVVVGRTAAPRPRPRRPPPLARLAAHT
Summary
Uniprot
H9JVJ3
A0A2A4JZ87
A0A2A4K117
A0A2H1WSD6
A0A212ESY7
A0A2A4K0P0
+ More
A0A194PLB6 A0A194RBX5 A0A0L7LAM0 A0A1B0CSP3 A0A1B6DYV1 R4WS06 B0WPG2 A0A026WA75 E2BUY3 A0A3Q0IV98 A0A1S4G219 A0A182GKE3 Q16GK5 E2ACQ7 A0A3L8DBS5 E9ISB7 A0A151WHQ5 A0A2A3EQ30 A0A195DMI9 A0A151ILG0 A0A195FKA1 A0A195AW45 A0A067RIL4 A0A088ACM0 A0A158P213 K7IVU7 A0A0C9R7G4 A0A139WHN9 A0A1Y1JVX6 A0A232ETS3 A0A182YMD5 A0A182MRA1 A0A034VKW1 A0A0K8W256 A0A224XHJ8 A0A2H8TTI8 A0A1I8NEA1 A0A139WHZ4 A0A182RCP1 W5JM89 A0A1I8PE72 A0A2M4CM71 A0A182W6X1 Q7Q434 A0A2S2QSD2 E0VJS6 A0A1A9WKW4 A0A182NFB5 Q8T0D8 A0A2M4A3T2 U4UGK7 A0A1S4GY82 A0A182J5W7 A0A182JTG9 W8BRC3 A0A182HUH9 A0A0L0C4T6 A0A182V1H9 A0A182PPP6 A0A1A9XCS6 A0A1B0C456 A0A1B0AUS3 A0A1A9ZD16 A0A1A9UL15 A0A1B0FBV3 T1HDA6 A0A336LUX4 Q29L38 B4H8S1 A0A3B0K1I3 A0A182QTX8 A0A182F274 X2J9V1 A0A182SX27 Q9VLT6 B4Q691 A0A1B6G8W8 A0A0J9QXT2 B4HYD9 B3N6Z6 B4NWC9 A0A1W4WJS8 A0A1W4WKR7 B3MMX6 N6UFE6 A0A1W4W7B5 A0A0K8SA37 A0A182L194 A0A182TW08 A0A182WTW3 A0A146L5D8 B4MUI0
A0A194PLB6 A0A194RBX5 A0A0L7LAM0 A0A1B0CSP3 A0A1B6DYV1 R4WS06 B0WPG2 A0A026WA75 E2BUY3 A0A3Q0IV98 A0A1S4G219 A0A182GKE3 Q16GK5 E2ACQ7 A0A3L8DBS5 E9ISB7 A0A151WHQ5 A0A2A3EQ30 A0A195DMI9 A0A151ILG0 A0A195FKA1 A0A195AW45 A0A067RIL4 A0A088ACM0 A0A158P213 K7IVU7 A0A0C9R7G4 A0A139WHN9 A0A1Y1JVX6 A0A232ETS3 A0A182YMD5 A0A182MRA1 A0A034VKW1 A0A0K8W256 A0A224XHJ8 A0A2H8TTI8 A0A1I8NEA1 A0A139WHZ4 A0A182RCP1 W5JM89 A0A1I8PE72 A0A2M4CM71 A0A182W6X1 Q7Q434 A0A2S2QSD2 E0VJS6 A0A1A9WKW4 A0A182NFB5 Q8T0D8 A0A2M4A3T2 U4UGK7 A0A1S4GY82 A0A182J5W7 A0A182JTG9 W8BRC3 A0A182HUH9 A0A0L0C4T6 A0A182V1H9 A0A182PPP6 A0A1A9XCS6 A0A1B0C456 A0A1B0AUS3 A0A1A9ZD16 A0A1A9UL15 A0A1B0FBV3 T1HDA6 A0A336LUX4 Q29L38 B4H8S1 A0A3B0K1I3 A0A182QTX8 A0A182F274 X2J9V1 A0A182SX27 Q9VLT6 B4Q691 A0A1B6G8W8 A0A0J9QXT2 B4HYD9 B3N6Z6 B4NWC9 A0A1W4WJS8 A0A1W4WKR7 B3MMX6 N6UFE6 A0A1W4W7B5 A0A0K8SA37 A0A182L194 A0A182TW08 A0A182WTW3 A0A146L5D8 B4MUI0
Pubmed
19121390
22118469
26354079
26227816
23691247
24508170
+ More
20798317 26483478 17510324 30249741 21282665 24845553 21347285 20075255 18362917 19820115 28004739 28648823 25244985 25348373 25315136 20920257 23761445 12364791 20566863 23537049 24495485 26108605 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 20966253 26823975
20798317 26483478 17510324 30249741 21282665 24845553 21347285 20075255 18362917 19820115 28004739 28648823 25244985 25348373 25315136 20920257 23761445 12364791 20566863 23537049 24495485 26108605 15632085 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 20966253 26823975
EMBL
BABH01038988
BABH01038989
BABH01038990
BABH01038991
BABH01038992
NWSH01000331
+ More
PCG77331.1 PCG77332.1 ODYU01010684 SOQ55958.1 AGBW02012699 OWR44600.1 PCG77333.1 KQ459601 KPI93788.1 KQ460398 KPJ15102.1 JTDY01002042 KOB72256.1 AJWK01026351 AJWK01026352 AJWK01026353 AJWK01026354 AJWK01026355 GEDC01006457 JAS30841.1 AK417482 BAN20697.1 DS232023 EDS32341.1 KK107310 EZA52878.1 GL450772 EFN80520.1 JXUM01069817 KQ562571 KXJ75580.1 CH478268 EAT33370.1 GL438568 EFN68801.1 QOIP01000010 RLU17960.1 GL765324 EFZ16551.1 KQ983112 KYQ47373.1 KZ288204 PBC33239.1 KQ980724 KYN14098.1 KQ977120 KYN05592.1 KQ981523 KYN40429.1 KQ976731 KYM76277.1 KK852451 KDR23676.1 ADTU01006956 GBYB01002135 GBYB01002136 GBYB01002137 GBYB01008737 JAG71902.1 JAG71903.1 JAG71904.1 JAG78504.1 KQ971343 KYB27399.1 GEZM01099122 JAV53482.1 NNAY01002229 OXU21760.1 AXCM01002055 GAKP01014996 JAC43956.1 GDHF01007135 JAI45179.1 GFTR01008915 JAW07511.1 GFXV01005760 MBW17565.1 KYB27397.1 ADMH02000711 ETN65266.1 GGFL01002248 MBW66426.1 AAAB01008964 EAA12463.4 GGMS01011217 MBY80420.1 DS235226 EEB13632.1 AY069385 AAL39530.1 GGFK01002098 MBW35419.1 KB632288 ERL91478.1 GAMC01010764 JAB95791.1 APCN01002461 JRES01000905 JRES01000841 KNC27368.1 KNC27827.1 JXJN01025273 JXJN01003724 CCAG010019860 ACPB03001477 ACPB03001478 UFQS01000215 UFQT01000215 SSX01456.1 SSX21836.1 CH379061 EAL32986.3 CH479225 EDW35130.1 OUUW01000004 SPP79819.1 AXCN02000822 AE014134 AHN54252.1 AAF52597.4 CM000361 EDX04167.1 GECZ01010889 JAS58880.1 CM002910 KMY88912.1 CH480818 EDW52069.1 CH954177 EDV59292.2 CM000157 EDW88446.2 CH902620 EDV31001.2 APGK01037607 APGK01037608 APGK01037609 APGK01037610 APGK01037611 APGK01037612 APGK01037613 KB740948 ENN77377.1 GBRD01015670 JAG50156.1 GDHC01016669 GDHC01015303 JAQ01960.1 JAQ03326.1 CH963857 EDW76175.2
PCG77331.1 PCG77332.1 ODYU01010684 SOQ55958.1 AGBW02012699 OWR44600.1 PCG77333.1 KQ459601 KPI93788.1 KQ460398 KPJ15102.1 JTDY01002042 KOB72256.1 AJWK01026351 AJWK01026352 AJWK01026353 AJWK01026354 AJWK01026355 GEDC01006457 JAS30841.1 AK417482 BAN20697.1 DS232023 EDS32341.1 KK107310 EZA52878.1 GL450772 EFN80520.1 JXUM01069817 KQ562571 KXJ75580.1 CH478268 EAT33370.1 GL438568 EFN68801.1 QOIP01000010 RLU17960.1 GL765324 EFZ16551.1 KQ983112 KYQ47373.1 KZ288204 PBC33239.1 KQ980724 KYN14098.1 KQ977120 KYN05592.1 KQ981523 KYN40429.1 KQ976731 KYM76277.1 KK852451 KDR23676.1 ADTU01006956 GBYB01002135 GBYB01002136 GBYB01002137 GBYB01008737 JAG71902.1 JAG71903.1 JAG71904.1 JAG78504.1 KQ971343 KYB27399.1 GEZM01099122 JAV53482.1 NNAY01002229 OXU21760.1 AXCM01002055 GAKP01014996 JAC43956.1 GDHF01007135 JAI45179.1 GFTR01008915 JAW07511.1 GFXV01005760 MBW17565.1 KYB27397.1 ADMH02000711 ETN65266.1 GGFL01002248 MBW66426.1 AAAB01008964 EAA12463.4 GGMS01011217 MBY80420.1 DS235226 EEB13632.1 AY069385 AAL39530.1 GGFK01002098 MBW35419.1 KB632288 ERL91478.1 GAMC01010764 JAB95791.1 APCN01002461 JRES01000905 JRES01000841 KNC27368.1 KNC27827.1 JXJN01025273 JXJN01003724 CCAG010019860 ACPB03001477 ACPB03001478 UFQS01000215 UFQT01000215 SSX01456.1 SSX21836.1 CH379061 EAL32986.3 CH479225 EDW35130.1 OUUW01000004 SPP79819.1 AXCN02000822 AE014134 AHN54252.1 AAF52597.4 CM000361 EDX04167.1 GECZ01010889 JAS58880.1 CM002910 KMY88912.1 CH480818 EDW52069.1 CH954177 EDV59292.2 CM000157 EDW88446.2 CH902620 EDV31001.2 APGK01037607 APGK01037608 APGK01037609 APGK01037610 APGK01037611 APGK01037612 APGK01037613 KB740948 ENN77377.1 GBRD01015670 JAG50156.1 GDHC01016669 GDHC01015303 JAQ01960.1 JAQ03326.1 CH963857 EDW76175.2
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000092461 UP000002320 UP000053097 UP000008237 UP000079169 UP000069940 UP000249989 UP000008820 UP000000311 UP000279307 UP000075809 UP000242457 UP000078492 UP000078542 UP000078541 UP000078540 UP000027135 UP000005203 UP000005205 UP000002358 UP000007266 UP000215335 UP000076408 UP000075883 UP000095301 UP000075900 UP000000673 UP000095300 UP000075920 UP000007062 UP000009046 UP000091820 UP000075884 UP000030742 UP000075880 UP000075881 UP000075840 UP000037069 UP000075903 UP000075885 UP000092443 UP000092460 UP000092445 UP000078200 UP000092444 UP000015103 UP000001819 UP000008744 UP000268350 UP000075886 UP000069272 UP000000803 UP000075901 UP000000304 UP000001292 UP000008711 UP000002282 UP000192223 UP000007801 UP000019118 UP000192221 UP000075882 UP000075902 UP000076407 UP000007798
UP000092461 UP000002320 UP000053097 UP000008237 UP000079169 UP000069940 UP000249989 UP000008820 UP000000311 UP000279307 UP000075809 UP000242457 UP000078492 UP000078542 UP000078541 UP000078540 UP000027135 UP000005203 UP000005205 UP000002358 UP000007266 UP000215335 UP000076408 UP000075883 UP000095301 UP000075900 UP000000673 UP000095300 UP000075920 UP000007062 UP000009046 UP000091820 UP000075884 UP000030742 UP000075880 UP000075881 UP000075840 UP000037069 UP000075903 UP000075885 UP000092443 UP000092460 UP000092445 UP000078200 UP000092444 UP000015103 UP000001819 UP000008744 UP000268350 UP000075886 UP000069272 UP000000803 UP000075901 UP000000304 UP000001292 UP000008711 UP000002282 UP000192223 UP000007801 UP000019118 UP000192221 UP000075882 UP000075902 UP000076407 UP000007798
Pfam
Interpro
IPR000152
EGF-type_Asp/Asn_hydroxyl_site
+ More
IPR000742 EGF-like_dom
IPR016201 PSI
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR002049 Laminin_EGF
IPR009030 Growth_fac_rcpt_cys_sf
IPR015915 Kelch-typ_b-propeller
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
IPR002165 Plexin_repeat
IPR001881 EGF-like_Ca-bd_dom
IPR011043 Gal_Oxase/kelch_b-propeller
IPR006652 Kelch_1
IPR001368 TNFR/NGFR_Cys_rich_reg
IPR008211 Laminin_N
IPR038684 Laminin_N_sf
IPR000033 LDLR_classB_rpt
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR011042 6-blade_b-propeller_TolB-like
IPR023415 LDLR_class-A_CS
IPR011047 Quinoprotein_ADH-like_supfam
IPR041161 EGF_Tenascin
IPR011498 Kelch_2
IPR000742 EGF-like_dom
IPR016201 PSI
IPR013032 EGF-like_CS
IPR018097 EGF_Ca-bd_CS
IPR002049 Laminin_EGF
IPR009030 Growth_fac_rcpt_cys_sf
IPR015915 Kelch-typ_b-propeller
IPR000859 CUB_dom
IPR035914 Sperma_CUB_dom_sf
IPR002165 Plexin_repeat
IPR001881 EGF-like_Ca-bd_dom
IPR011043 Gal_Oxase/kelch_b-propeller
IPR006652 Kelch_1
IPR001368 TNFR/NGFR_Cys_rich_reg
IPR008211 Laminin_N
IPR038684 Laminin_N_sf
IPR000033 LDLR_classB_rpt
IPR002172 LDrepeatLR_classA_rpt
IPR036055 LDL_receptor-like_sf
IPR011042 6-blade_b-propeller_TolB-like
IPR023415 LDLR_class-A_CS
IPR011047 Quinoprotein_ADH-like_supfam
IPR041161 EGF_Tenascin
IPR011498 Kelch_2
SUPFAM
ProteinModelPortal
H9JVJ3
A0A2A4JZ87
A0A2A4K117
A0A2H1WSD6
A0A212ESY7
A0A2A4K0P0
+ More
A0A194PLB6 A0A194RBX5 A0A0L7LAM0 A0A1B0CSP3 A0A1B6DYV1 R4WS06 B0WPG2 A0A026WA75 E2BUY3 A0A3Q0IV98 A0A1S4G219 A0A182GKE3 Q16GK5 E2ACQ7 A0A3L8DBS5 E9ISB7 A0A151WHQ5 A0A2A3EQ30 A0A195DMI9 A0A151ILG0 A0A195FKA1 A0A195AW45 A0A067RIL4 A0A088ACM0 A0A158P213 K7IVU7 A0A0C9R7G4 A0A139WHN9 A0A1Y1JVX6 A0A232ETS3 A0A182YMD5 A0A182MRA1 A0A034VKW1 A0A0K8W256 A0A224XHJ8 A0A2H8TTI8 A0A1I8NEA1 A0A139WHZ4 A0A182RCP1 W5JM89 A0A1I8PE72 A0A2M4CM71 A0A182W6X1 Q7Q434 A0A2S2QSD2 E0VJS6 A0A1A9WKW4 A0A182NFB5 Q8T0D8 A0A2M4A3T2 U4UGK7 A0A1S4GY82 A0A182J5W7 A0A182JTG9 W8BRC3 A0A182HUH9 A0A0L0C4T6 A0A182V1H9 A0A182PPP6 A0A1A9XCS6 A0A1B0C456 A0A1B0AUS3 A0A1A9ZD16 A0A1A9UL15 A0A1B0FBV3 T1HDA6 A0A336LUX4 Q29L38 B4H8S1 A0A3B0K1I3 A0A182QTX8 A0A182F274 X2J9V1 A0A182SX27 Q9VLT6 B4Q691 A0A1B6G8W8 A0A0J9QXT2 B4HYD9 B3N6Z6 B4NWC9 A0A1W4WJS8 A0A1W4WKR7 B3MMX6 N6UFE6 A0A1W4W7B5 A0A0K8SA37 A0A182L194 A0A182TW08 A0A182WTW3 A0A146L5D8 B4MUI0
A0A194PLB6 A0A194RBX5 A0A0L7LAM0 A0A1B0CSP3 A0A1B6DYV1 R4WS06 B0WPG2 A0A026WA75 E2BUY3 A0A3Q0IV98 A0A1S4G219 A0A182GKE3 Q16GK5 E2ACQ7 A0A3L8DBS5 E9ISB7 A0A151WHQ5 A0A2A3EQ30 A0A195DMI9 A0A151ILG0 A0A195FKA1 A0A195AW45 A0A067RIL4 A0A088ACM0 A0A158P213 K7IVU7 A0A0C9R7G4 A0A139WHN9 A0A1Y1JVX6 A0A232ETS3 A0A182YMD5 A0A182MRA1 A0A034VKW1 A0A0K8W256 A0A224XHJ8 A0A2H8TTI8 A0A1I8NEA1 A0A139WHZ4 A0A182RCP1 W5JM89 A0A1I8PE72 A0A2M4CM71 A0A182W6X1 Q7Q434 A0A2S2QSD2 E0VJS6 A0A1A9WKW4 A0A182NFB5 Q8T0D8 A0A2M4A3T2 U4UGK7 A0A1S4GY82 A0A182J5W7 A0A182JTG9 W8BRC3 A0A182HUH9 A0A0L0C4T6 A0A182V1H9 A0A182PPP6 A0A1A9XCS6 A0A1B0C456 A0A1B0AUS3 A0A1A9ZD16 A0A1A9UL15 A0A1B0FBV3 T1HDA6 A0A336LUX4 Q29L38 B4H8S1 A0A3B0K1I3 A0A182QTX8 A0A182F274 X2J9V1 A0A182SX27 Q9VLT6 B4Q691 A0A1B6G8W8 A0A0J9QXT2 B4HYD9 B3N6Z6 B4NWC9 A0A1W4WJS8 A0A1W4WKR7 B3MMX6 N6UFE6 A0A1W4W7B5 A0A0K8SA37 A0A182L194 A0A182TW08 A0A182WTW3 A0A146L5D8 B4MUI0
PDB
6FKQ
E-value=0.000113272,
Score=109
Ontologies
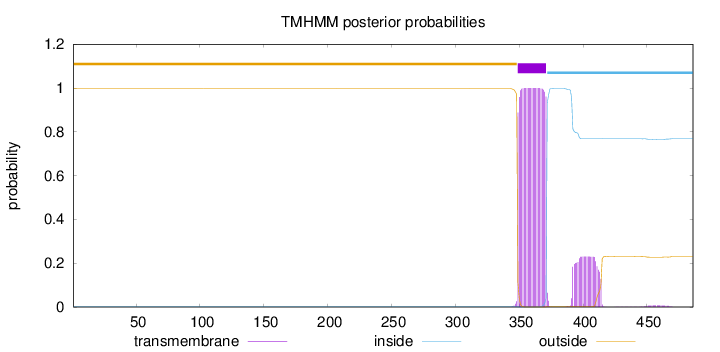
Topology
Length:
486
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
27.92806
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00277
outside
1 - 348
TMhelix
349 - 371
inside
372 - 486
Population Genetic Test Statistics
Pi
234.267041
Theta
162.544567
Tajima's D
1.356501
CLR
0.584571
CSRT
0.754412279386031
Interpretation
Uncertain
