Pre Gene Modal
BGIBMGA004307
Annotation
PREDICTED:_enhancer_of_yellow_2_transcription_factor-like_[Amyelois_transitella]
Full name
Enhancer of yellow 2 transcription factor
+ More
Transcription and mRNA export factor ENY2
Transcription and mRNA export factor ENY2
Alternative Name
Enhancer of yellow 2 transcription factor homolog
Location in the cell
Mitochondrial Reliability : 1.592 Nuclear Reliability : 1.947
Sequence
CDS
ATGACTGTGAATAACACCATTGCACACCAACGTTTAATTCTTAGTGGAGATCGAGAAAGGTTTAAGGAGCTTTTGAGAAGGAGATTAATAGAATGTGGATGGAGAGATCAAGTCAGAATGCTATGTCGAGACATAGTGAAAGAAAATGAGAGCGGAAACATCACATTTGATTCTTTAGTAACAAAGGTGACTCCCCGTGCTAGAGCACTTGTGCCAGATTCAGTGAAAAAAGAATTATTGCAGAAAATTAAAACCCATCTACTCACACAGAAAGACCAATAA
Protein
MTVNNTIAHQRLILSGDRERFKELLRRRLIECGWRDQVRMLCRDIVKENESGNITFDSLVTKVTPRARALVPDSVKKELLQKIKTHLLTQKDQ
Summary
Description
Involved in mRNA export coupled transcription activation by association with both the TREX-2 and the SAGA complexes. The transcription regulatory histone acetylation (HAT) complex SAGA is a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates to a subcomplex that specifically deubiquitinates histones. The SAGA complex is recruited to specific gene promoters by activators, where it is required for transcription. The TREX-2 complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). TREX-2 participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery.
Involved in mRNA export coupled transcription activation by association with both the AMEX and the SAGA complexes. The SAGA complex is a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates in a subcomplex that specifically deubiquitinates histone H2B. The SAGA complex is recruited to specific gene promoters by activators, where it is required for transcription. Required for nuclear receptor-mediated transactivation. Involved in transcription elongation by recruiting the THO complex onto nascent mRNA. The AMEX complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). AMEX participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery (By similarity).
Involved in mRNA export coupled transcription activation by association with both the TREX-2 and the SAGA complexes. The transcription regulatory histone acetylation (HAT) complex SAGA is a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates in a subcomplex that specifically deubiquitinates histones. The SAGA complex is recruited to specific gene promoters by activators, where it is required for transcription. The TREX-2 complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). TREX-2 participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery (By similarity).
Involved in mRNA export coupled transcription activation by association with both the TREX-2 and the SAGA complexes. The transcription regulatory histone acetylation (HAT) complex SAGA is a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates to a subcomplex that specifically deubiquitinates both histones H2A and H2B. The SAGA complex is recruited to specific gene promoters by activators such as MYC, where it is required for transcription. Required for nuclear receptor-mediated transactivation. The TREX-2 complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). TREX-2 participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery.
Involved in mRNA export coupled transcription activation by association with both the AMEX and the SAGA complexes. The SAGA complex is a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates to a subcomplex that specifically deubiquitinates histone H2B. The SAGA complex is recruited to specific gene promoters by activators, where it is required for transcription. Required for nuclear receptor-mediated transactivation. Involved in transcription elongation by recruiting the THO complex onto nascent mRNA. The AMEX complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). AMEX participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery.
Involved in mRNA export coupled transcription activation by association with both the AMEX and the SAGA complexes. The SAGA complex is a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates in a subcomplex that specifically deubiquitinates histone H2B. The SAGA complex is recruited to specific gene promoters by activators, where it is required for transcription. Required for nuclear receptor-mediated transactivation. Involved in transcription elongation by recruiting the THO complex onto nascent mRNA. The AMEX complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). AMEX participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery (By similarity).
Involved in mRNA export coupled transcription activation by association with both the TREX-2 and the SAGA complexes. The transcription regulatory histone acetylation (HAT) complex SAGA is a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates in a subcomplex that specifically deubiquitinates histones. The SAGA complex is recruited to specific gene promoters by activators, where it is required for transcription. The TREX-2 complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). TREX-2 participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery (By similarity).
Involved in mRNA export coupled transcription activation by association with both the TREX-2 and the SAGA complexes. The transcription regulatory histone acetylation (HAT) complex SAGA is a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates to a subcomplex that specifically deubiquitinates both histones H2A and H2B. The SAGA complex is recruited to specific gene promoters by activators such as MYC, where it is required for transcription. Required for nuclear receptor-mediated transactivation. The TREX-2 complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). TREX-2 participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery.
Involved in mRNA export coupled transcription activation by association with both the AMEX and the SAGA complexes. The SAGA complex is a multiprotein complex that activates transcription by remodeling chromatin and mediating histone acetylation and deubiquitination. Within the SAGA complex, participates to a subcomplex that specifically deubiquitinates histone H2B. The SAGA complex is recruited to specific gene promoters by activators, where it is required for transcription. Required for nuclear receptor-mediated transactivation. Involved in transcription elongation by recruiting the THO complex onto nascent mRNA. The AMEX complex functions in docking export-competent ribonucleoprotein particles (mRNPs) to the nuclear entrance of the nuclear pore complex (nuclear basket). AMEX participates in mRNA export and accurate chromatin positioning in the nucleus by tethering genes to the nuclear periphery.
Subunit
Component of the nuclear pore complex (NPC)-associated TREX-2 complex (transcription and export complex 2). Component of the SAGA transcription coactivator-HAT complex. Within the SAGA complex, participates to a subcomplex of SAGA called the DUB module (deubiquitination module).
Component of the nuclear pore complex (NPC)-associated AMEX complex (anchoring and mRNA export complex), composed of at least e(y)2 and xmas-2. Component of the SAGA transcription coactivator-HAT complexes, at least composed of Ada2b, e(y)2, Pcaf/Gcn5, Taf10 and Nipped-A/Trrap. Within the SAGA complex, e(y)2, Sgf11, and not/nonstop form an additional subcomplex of SAGA called the DUB module (deubiquitination module). Component of the THO complex, composed of at least e(y)2, HPR1, THO2, THOC5, THOC6 and THOC7. Interacts with e(y)1. Interacts with su(Hw) (via zinc fingers). Interacts with xmas-2; required for localization to the nuclear periphery. Interacts with the nuclear pore complex (NPC).
Component of the nuclear pore complex (NPC)-associated TREX-2 complex (transcription and export complex 2). Component of the SAGA transcription coactivator-HAT complex. Within the SAGA complex, participates in a subcomplex of SAGA called the DUB module (deubiquitination module) (By similarity).
Component of the nuclear pore complex (NPC)-associated TREX-2 complex (transcription and export complex 2), composed of at least ENY2, GANP, PCID2, DSS1, and either centrin CETN2 or CETN3. TREX-2 contains 2 ENY2 chains. The TREX-2 complex interacts with the nucleoporin NUP153. Component of some SAGA transcription coactivator-HAT complexes, at least composed of ATXN7, ATXN7L3, ENY2, GCN5L2, SUPT3H, TAF10, TRRAP and USP22. Within the SAGA complex, ENY2, ATXN7, ATXN7L3, and USP22 form an additional subcomplex of SAGA called the DUB module (deubiquitination module). Interacts directly with ATXN7L3, GANP and with the RNA polymerase II. Interacts strongly with ATXN7L3 and ATXN7L3B.
Component of the nuclear pore complex (NPC)-associated AMEX complex (anchoring and mRNA export complex), composed of at least e(y)2 and xmas-2. Component of the SAGA transcription coactivator-HAT complexes, at least composed of Ada2b, e(y)2, Pcaf/Gcn5, Taf10 and Nipped-A/Trrap. Within the SAGA complex, e(y)2, Sgf11, and not/nonstop form an additional subcomplex of SAGA called the DUB module (deubiquitination module). Component of the THO complex, composed of at least e(y)2, HPR1, THO2, THOC5, THOC6 and THOC7. Interacts with e(y)1. Interacts with su(Hw) (via zinc fingers). Interacts with xmas-2; required for localization to the nuclear periphery. Interacts with the nuclear pore complex (NPC).
Component of the nuclear pore complex (NPC)-associated TREX-2 complex (transcription and export complex 2). Component of the SAGA transcription coactivator-HAT complex. Within the SAGA complex, participates in a subcomplex of SAGA called the DUB module (deubiquitination module) (By similarity).
Component of the nuclear pore complex (NPC)-associated TREX-2 complex (transcription and export complex 2), composed of at least ENY2, GANP, PCID2, DSS1, and either centrin CETN2 or CETN3. TREX-2 contains 2 ENY2 chains. The TREX-2 complex interacts with the nucleoporin NUP153. Component of some SAGA transcription coactivator-HAT complexes, at least composed of ATXN7, ATXN7L3, ENY2, GCN5L2, SUPT3H, TAF10, TRRAP and USP22. Within the SAGA complex, ENY2, ATXN7, ATXN7L3, and USP22 form an additional subcomplex of SAGA called the DUB module (deubiquitination module). Interacts directly with ATXN7L3, GANP and with the RNA polymerase II. Interacts strongly with ATXN7L3 and ATXN7L3B.
Similarity
Belongs to the ENY2 family.
Keywords
Activator
Chromatin regulator
Complete proteome
Cytoplasm
mRNA transport
Nuclear pore complex
Nucleus
Protein transport
Reference proteome
Transcription
Transcription regulation
Translocation
Transport
Feature
chain Enhancer of yellow 2 transcription factor
Uniprot
A0A2A4JZU5
A0A2A4K0Y0
A0A2H1WSA5
A0A194RBA0
A0A212ESZ9
I4DKX2
+ More
S4NVK7 H9J468 A0A026WV43 F4WGD0 A0A158NI46 A0A2A3E8Y5 A0A088A320 A0A310SHK6 A0A0L7QXG2 A0A0M9A7G2 A0A0J7KHT3 A0A2P8YTI2 B3MQ24 A0A195CP71 A0A154P5B9 B4M6M6 A0A195F069 A0A067RKG9 A0A195EFZ5 E2AUI3 A0A151WJH2 A0A023EEE4 Q17MZ8 E2BV72 A0A232FDK3 K7J8Z4 T1ISW1 B4N1G8 A0A0L0BWC3 B4L1Z8 T1FHE2 A0A1I8MGG6 A0A1Q3F4P9 K4G0B6 A0A1B0FLE0 A0A1B0BSM9 A0A1A9Z3N5 A0A1Q3F471 F7G3F4 Q6DH42 A0A0P8YST3 A0A1B0EU97 B0WG73 A0A093IXG8 B4JLC3 C3ZZM0 A0A1Z5KXE8 F7C227 A0A091G502 A0A091FWX7 G3VFQ5 A0A2I0UU06 A0A3L8T0P2 A0A218VDK2 G1KYQ7 A0A0B8RRU7 U3JUD0 A0A0Q3XAL6 V8NTG8 A0A2I0MWF1 A0A098LYV6 B5FZ63 A0A091HE87 A0A093Q831 A0A091HPW1 A0A091UKD0 A0A091J791 A0A099ZYG2 A0A091QWC4 A0A093NBA4 A0A091SII3 A0A087R772 H0ZPT2 A0A091R1S9 A0A099ZVZ9 A0A093H7H5 A0A091VFN9 A0A1V4KH29 A0A087VBV9 A0A1I8PE17 A0A1B1JCE7 A0A3N0XS03 K7FHN5 A0A094K7S1 A0A1D5PYW8 U5EXI7 R0LHK8 A0A1S3IAJ2 A0A1W5AZL6 H3AXA7 Q29IN4 A0A1L8FZW0 Q3KPT5
S4NVK7 H9J468 A0A026WV43 F4WGD0 A0A158NI46 A0A2A3E8Y5 A0A088A320 A0A310SHK6 A0A0L7QXG2 A0A0M9A7G2 A0A0J7KHT3 A0A2P8YTI2 B3MQ24 A0A195CP71 A0A154P5B9 B4M6M6 A0A195F069 A0A067RKG9 A0A195EFZ5 E2AUI3 A0A151WJH2 A0A023EEE4 Q17MZ8 E2BV72 A0A232FDK3 K7J8Z4 T1ISW1 B4N1G8 A0A0L0BWC3 B4L1Z8 T1FHE2 A0A1I8MGG6 A0A1Q3F4P9 K4G0B6 A0A1B0FLE0 A0A1B0BSM9 A0A1A9Z3N5 A0A1Q3F471 F7G3F4 Q6DH42 A0A0P8YST3 A0A1B0EU97 B0WG73 A0A093IXG8 B4JLC3 C3ZZM0 A0A1Z5KXE8 F7C227 A0A091G502 A0A091FWX7 G3VFQ5 A0A2I0UU06 A0A3L8T0P2 A0A218VDK2 G1KYQ7 A0A0B8RRU7 U3JUD0 A0A0Q3XAL6 V8NTG8 A0A2I0MWF1 A0A098LYV6 B5FZ63 A0A091HE87 A0A093Q831 A0A091HPW1 A0A091UKD0 A0A091J791 A0A099ZYG2 A0A091QWC4 A0A093NBA4 A0A091SII3 A0A087R772 H0ZPT2 A0A091R1S9 A0A099ZVZ9 A0A093H7H5 A0A091VFN9 A0A1V4KH29 A0A087VBV9 A0A1I8PE17 A0A1B1JCE7 A0A3N0XS03 K7FHN5 A0A094K7S1 A0A1D5PYW8 U5EXI7 R0LHK8 A0A1S3IAJ2 A0A1W5AZL6 H3AXA7 Q29IN4 A0A1L8FZW0 Q3KPT5
Pubmed
26354079
22118469
22651552
23622113
19121390
24508170
+ More
30249741 21719571 21347285 29403074 17994087 24845553 20798317 24945155 26483478 17510324 28648823 20075255 26108605 23254933 25315136 23056606 18464734 18563158 28528879 17495919 21709235 30282656 25476704 24297900 23371554 17018643 20360741 18574105 17381049 15592404 9215903 15632085 27762356
30249741 21719571 21347285 29403074 17994087 24845553 20798317 24945155 26483478 17510324 28648823 20075255 26108605 23254933 25315136 23056606 18464734 18563158 28528879 17495919 21709235 30282656 25476704 24297900 23371554 17018643 20360741 18574105 17381049 15592404 9215903 15632085 27762356
EMBL
NWSH01000331
PCG77329.1
PCG77330.1
ODYU01010684
SOQ55959.1
KQ460398
+ More
KPJ15103.1 AGBW02012699 OWR44599.1 AK401940 KQ459601 BAM18562.1 KPI93789.1 GAIX01012912 JAA79648.1 BABH01032542 KK107111 QOIP01000011 EZA58974.1 RLU17169.1 GL888128 EGI66897.1 ADTU01016271 KZ288322 PBC28185.1 KQ761039 OAD58350.1 KQ414704 KOC63295.1 KQ435719 KOX78767.1 LBMM01007353 KMQ89812.1 PYGN01000369 PSN47545.1 CH902621 KQ977481 KYN02440.1 KQ434809 KZC06378.1 CH940653 KQ981897 KYN33751.1 KK852415 KDR24366.1 KQ978957 KYN27086.1 GL442821 EFN62909.1 KQ983039 KYQ48008.1 JXUM01049131 JXUM01132269 GAPW01006594 GAPW01006593 KQ567821 KQ561591 JAC07005.1 KXJ69262.1 KXJ78081.1 CH477203 GL450824 EFN80364.1 NNAY01000412 OXU28590.1 JH431448 CH963925 JRES01001243 KNC24323.1 CH933810 AMQM01007834 KB097673 ESN92064.1 GFDL01012587 JAV22458.1 JX052753 AFK10981.1 CCAG010010324 JXJN01019798 GFDL01012691 JAV22354.1 AAPN01402473 BC076140 KPU81606.1 AJWK01014147 DS231923 KL216808 KFV71341.1 CH916370 GG666758 EEN42010.1 GFJQ02007265 JAV99704.1 KL447782 KFO77008.1 KK719751 KFO65880.1 AEFK01066052 KZ505636 PKU49541.1 QUSF01000001 RLW13267.1 MUZQ01000006 OWK64043.1 GBSH01000970 JAG68055.1 AGTO01010677 LMAW01000115 KQL60588.1 AZIM01002083 ETE64842.1 AKCR02000001 PKK34003.1 GBSI01000824 JAC95672.1 DQ214330 KL534109 KFO93784.1 KL670890 KFW80357.1 KL217684 KFO97876.1 KL409711 KFQ91359.1 KK501492 KFP15595.1 KL870020 KGL86747.1 KK683590 KFQ13649.1 KL224593 KFW61466.1 KK473128 KFQ58148.1 KL226153 KFM09326.1 ABQF01006042 KK710805 KFQ33194.1 KL897924 KGL84965.1 KL205974 KFV77626.1 KK733943 KFR01570.1 LSYS01003169 OPJ83776.1 KL487555 KFO10101.1 KU359107 ANS11586.1 RJVU01062584 ROJ29348.1 AGCU01085412 AGCU01085413 KL344018 KFZ54684.1 AADN05000663 GANO01000932 JAB58939.1 KB743132 EOB01110.1 AFYH01148797 AFYH01148798 AFYH01148799 AFYH01148800 CH379063 CM004476 OCT77097.1 BC106564
KPJ15103.1 AGBW02012699 OWR44599.1 AK401940 KQ459601 BAM18562.1 KPI93789.1 GAIX01012912 JAA79648.1 BABH01032542 KK107111 QOIP01000011 EZA58974.1 RLU17169.1 GL888128 EGI66897.1 ADTU01016271 KZ288322 PBC28185.1 KQ761039 OAD58350.1 KQ414704 KOC63295.1 KQ435719 KOX78767.1 LBMM01007353 KMQ89812.1 PYGN01000369 PSN47545.1 CH902621 KQ977481 KYN02440.1 KQ434809 KZC06378.1 CH940653 KQ981897 KYN33751.1 KK852415 KDR24366.1 KQ978957 KYN27086.1 GL442821 EFN62909.1 KQ983039 KYQ48008.1 JXUM01049131 JXUM01132269 GAPW01006594 GAPW01006593 KQ567821 KQ561591 JAC07005.1 KXJ69262.1 KXJ78081.1 CH477203 GL450824 EFN80364.1 NNAY01000412 OXU28590.1 JH431448 CH963925 JRES01001243 KNC24323.1 CH933810 AMQM01007834 KB097673 ESN92064.1 GFDL01012587 JAV22458.1 JX052753 AFK10981.1 CCAG010010324 JXJN01019798 GFDL01012691 JAV22354.1 AAPN01402473 BC076140 KPU81606.1 AJWK01014147 DS231923 KL216808 KFV71341.1 CH916370 GG666758 EEN42010.1 GFJQ02007265 JAV99704.1 KL447782 KFO77008.1 KK719751 KFO65880.1 AEFK01066052 KZ505636 PKU49541.1 QUSF01000001 RLW13267.1 MUZQ01000006 OWK64043.1 GBSH01000970 JAG68055.1 AGTO01010677 LMAW01000115 KQL60588.1 AZIM01002083 ETE64842.1 AKCR02000001 PKK34003.1 GBSI01000824 JAC95672.1 DQ214330 KL534109 KFO93784.1 KL670890 KFW80357.1 KL217684 KFO97876.1 KL409711 KFQ91359.1 KK501492 KFP15595.1 KL870020 KGL86747.1 KK683590 KFQ13649.1 KL224593 KFW61466.1 KK473128 KFQ58148.1 KL226153 KFM09326.1 ABQF01006042 KK710805 KFQ33194.1 KL897924 KGL84965.1 KL205974 KFV77626.1 KK733943 KFR01570.1 LSYS01003169 OPJ83776.1 KL487555 KFO10101.1 KU359107 ANS11586.1 RJVU01062584 ROJ29348.1 AGCU01085412 AGCU01085413 KL344018 KFZ54684.1 AADN05000663 GANO01000932 JAB58939.1 KB743132 EOB01110.1 AFYH01148797 AFYH01148798 AFYH01148799 AFYH01148800 CH379063 CM004476 OCT77097.1 BC106564
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000005204
UP000053097
+ More
UP000279307 UP000007755 UP000005205 UP000242457 UP000005203 UP000053825 UP000053105 UP000036403 UP000245037 UP000007801 UP000078542 UP000076502 UP000008792 UP000078541 UP000027135 UP000078492 UP000000311 UP000075809 UP000069940 UP000249989 UP000008820 UP000008237 UP000215335 UP000002358 UP000007798 UP000037069 UP000009192 UP000015101 UP000095301 UP000092444 UP000092460 UP000092445 UP000002279 UP000000437 UP000092461 UP000002320 UP000053875 UP000001070 UP000001554 UP000002280 UP000053760 UP000052976 UP000007648 UP000276834 UP000197619 UP000001646 UP000016665 UP000051836 UP000053872 UP000007754 UP000053258 UP000054308 UP000053283 UP000053119 UP000053858 UP000054081 UP000053286 UP000053641 UP000053584 UP000053605 UP000190648 UP000095300 UP000007267 UP000000539 UP000085678 UP000192224 UP000008672 UP000001819 UP000186698
UP000279307 UP000007755 UP000005205 UP000242457 UP000005203 UP000053825 UP000053105 UP000036403 UP000245037 UP000007801 UP000078542 UP000076502 UP000008792 UP000078541 UP000027135 UP000078492 UP000000311 UP000075809 UP000069940 UP000249989 UP000008820 UP000008237 UP000215335 UP000002358 UP000007798 UP000037069 UP000009192 UP000015101 UP000095301 UP000092444 UP000092460 UP000092445 UP000002279 UP000000437 UP000092461 UP000002320 UP000053875 UP000001070 UP000001554 UP000002280 UP000053760 UP000052976 UP000007648 UP000276834 UP000197619 UP000001646 UP000016665 UP000051836 UP000053872 UP000007754 UP000053258 UP000054308 UP000053283 UP000053119 UP000053858 UP000054081 UP000053286 UP000053641 UP000053584 UP000053605 UP000190648 UP000095300 UP000007267 UP000000539 UP000085678 UP000192224 UP000008672 UP000001819 UP000186698
Interpro
SUPFAM
SSF54236
SSF54236
Gene 3D
ProteinModelPortal
A0A2A4JZU5
A0A2A4K0Y0
A0A2H1WSA5
A0A194RBA0
A0A212ESZ9
I4DKX2
+ More
S4NVK7 H9J468 A0A026WV43 F4WGD0 A0A158NI46 A0A2A3E8Y5 A0A088A320 A0A310SHK6 A0A0L7QXG2 A0A0M9A7G2 A0A0J7KHT3 A0A2P8YTI2 B3MQ24 A0A195CP71 A0A154P5B9 B4M6M6 A0A195F069 A0A067RKG9 A0A195EFZ5 E2AUI3 A0A151WJH2 A0A023EEE4 Q17MZ8 E2BV72 A0A232FDK3 K7J8Z4 T1ISW1 B4N1G8 A0A0L0BWC3 B4L1Z8 T1FHE2 A0A1I8MGG6 A0A1Q3F4P9 K4G0B6 A0A1B0FLE0 A0A1B0BSM9 A0A1A9Z3N5 A0A1Q3F471 F7G3F4 Q6DH42 A0A0P8YST3 A0A1B0EU97 B0WG73 A0A093IXG8 B4JLC3 C3ZZM0 A0A1Z5KXE8 F7C227 A0A091G502 A0A091FWX7 G3VFQ5 A0A2I0UU06 A0A3L8T0P2 A0A218VDK2 G1KYQ7 A0A0B8RRU7 U3JUD0 A0A0Q3XAL6 V8NTG8 A0A2I0MWF1 A0A098LYV6 B5FZ63 A0A091HE87 A0A093Q831 A0A091HPW1 A0A091UKD0 A0A091J791 A0A099ZYG2 A0A091QWC4 A0A093NBA4 A0A091SII3 A0A087R772 H0ZPT2 A0A091R1S9 A0A099ZVZ9 A0A093H7H5 A0A091VFN9 A0A1V4KH29 A0A087VBV9 A0A1I8PE17 A0A1B1JCE7 A0A3N0XS03 K7FHN5 A0A094K7S1 A0A1D5PYW8 U5EXI7 R0LHK8 A0A1S3IAJ2 A0A1W5AZL6 H3AXA7 Q29IN4 A0A1L8FZW0 Q3KPT5
S4NVK7 H9J468 A0A026WV43 F4WGD0 A0A158NI46 A0A2A3E8Y5 A0A088A320 A0A310SHK6 A0A0L7QXG2 A0A0M9A7G2 A0A0J7KHT3 A0A2P8YTI2 B3MQ24 A0A195CP71 A0A154P5B9 B4M6M6 A0A195F069 A0A067RKG9 A0A195EFZ5 E2AUI3 A0A151WJH2 A0A023EEE4 Q17MZ8 E2BV72 A0A232FDK3 K7J8Z4 T1ISW1 B4N1G8 A0A0L0BWC3 B4L1Z8 T1FHE2 A0A1I8MGG6 A0A1Q3F4P9 K4G0B6 A0A1B0FLE0 A0A1B0BSM9 A0A1A9Z3N5 A0A1Q3F471 F7G3F4 Q6DH42 A0A0P8YST3 A0A1B0EU97 B0WG73 A0A093IXG8 B4JLC3 C3ZZM0 A0A1Z5KXE8 F7C227 A0A091G502 A0A091FWX7 G3VFQ5 A0A2I0UU06 A0A3L8T0P2 A0A218VDK2 G1KYQ7 A0A0B8RRU7 U3JUD0 A0A0Q3XAL6 V8NTG8 A0A2I0MWF1 A0A098LYV6 B5FZ63 A0A091HE87 A0A093Q831 A0A091HPW1 A0A091UKD0 A0A091J791 A0A099ZYG2 A0A091QWC4 A0A093NBA4 A0A091SII3 A0A087R772 H0ZPT2 A0A091R1S9 A0A099ZVZ9 A0A093H7H5 A0A091VFN9 A0A1V4KH29 A0A087VBV9 A0A1I8PE17 A0A1B1JCE7 A0A3N0XS03 K7FHN5 A0A094K7S1 A0A1D5PYW8 U5EXI7 R0LHK8 A0A1S3IAJ2 A0A1W5AZL6 H3AXA7 Q29IN4 A0A1L8FZW0 Q3KPT5
PDB
4DHX
E-value=1.70144e-13,
Score=177
Ontologies
GO
GO:0071819
GO:0006406
GO:0000124
GO:0006368
GO:0005643
GO:0003713
GO:0045893
GO:0016578
GO:0070390
GO:0034399
GO:0070742
GO:0001094
GO:0043035
GO:0016973
GO:0045944
GO:0005737
GO:0031936
GO:0000785
GO:0030374
GO:0016021
GO:0006357
GO:0003682
GO:0044615
GO:0005739
GO:0016876
GO:0043039
GO:0004651
GO:0005634
PANTHER
Topology
Subcellular location
Nucleus
Nucleoplasm
Nuclear pore complex
Cytoplasm
Nucleoplasm
Nuclear pore complex
Cytoplasm
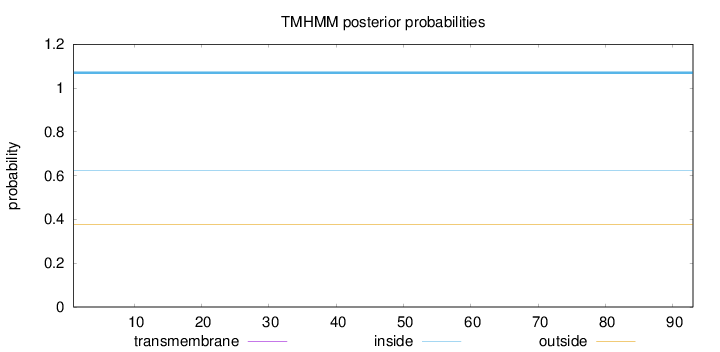
Length:
93
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.62196
inside
1 - 93
Population Genetic Test Statistics
Pi
435.569125
Theta
221.671891
Tajima's D
3.061713
CLR
0
CSRT
0.981150942452877
Interpretation
Uncertain
