Gene
KWMTBOMO01998
Pre Gene Modal
BGIBMGA013557
Annotation
PREDICTED:_peptidyl-prolyl_cis-trans_isomerase?_rhodopsin-specific_isozyme_isoform_X2_[Papilio_xuthus]
Full name
Peptidyl-prolyl cis-trans isomerase
+ More
Peptidyl-prolyl cis-trans isomerase, rhodopsin-specific isozyme
Peptidyl-prolyl cis-trans isomerase, rhodopsin-specific isozyme
Alternative Name
Rotamase
Location in the cell
Cytoplasmic Reliability : 1.433 Extracellular Reliability : 1.176
Sequence
CDS
ATGAAGTTTCTACAAGTTCTTTATACAGTGATATACTTTTCTTTACTTTTAAATCTTGTAGACGGACGACAATTTCGTGTTACTGATCAAGTGTACTTAGATGTTAAGAATGAAGACAAGGACCTGGGAAGACTTGTTATTGGACTGTTCGGAGATTTAGCTCCAAAAGCGGTGAATAATTTCAAAGTTCTCGCTACTAAAGGCATAAATGGGAAATCGTATAAAGGAACTCGGTTTAATCGTATCATCAAAAGGTTTATGGTCCAAGGCGGTGACGTCGTCTCCGATGACGGTAGTGGTAGCATTAGTATTTATGGAGATACATTTGACGATGAAAATCTGGAAACCCAGCATAGTACAGGAGGATTTGTGTCAATGGCCAATAAAGGTAAAAACACAAATGGCTGTCAGTTTTTGATAACCACCACTGGTGCGCCTTGGTTAGATCAACTGCACACTGTAATTGGGAAAGTCGTAGAAGGTCAGAAAGTGGTGCATCTGTTGGAGCACACGCCAACCGATGTTGAGGATCGACCGTTGGAGCATGTATATATTTCAGACTGTGGTCTGCTGCCGACACCGCAACCTTACTATATCTCCGACGATCCATACGATTTGTGGGCGTGGATCAAGGCCTCTGCAGTGCCGCTGACGATGTCATTTTCGATACTTGGTTTCTTTCATTGGATGATTAGGAAAATGGAAATTTAA
Protein
MKFLQVLYTVIYFSLLLNLVDGRQFRVTDQVYLDVKNEDKDLGRLVIGLFGDLAPKAVNNFKVLATKGINGKSYKGTRFNRIIKRFMVQGGDVVSDDGSGSISIYGDTFDDENLETQHSTGGFVSMANKGKNTNGCQFLITTTGAPWLDQLHTVIGKVVEGQKVVHLLEHTPTDVEDRPLEHVYISDCGLLPTPQPYYISDDPYDLWAWIKASAVPLTMSFSILGFFHWMIRKMEI
Summary
Description
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides.
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides. Acts on the folding of rhodopsin RH1 and RH2 (but not RH3) and is required for visual transduction.
PPIases accelerate the folding of proteins. It catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides. Acts on the folding of rhodopsin RH1 and RH2 (but not RH3) and is required for visual transduction.
Catalytic Activity
[protein]-peptidylproline (omega=180) = [protein]-peptidylproline (omega=0)
Similarity
Belongs to the cyclophilin-type PPIase family.
Keywords
Complete proteome
Glycoprotein
Isomerase
Membrane
Reference proteome
Rotamase
Sensory transduction
Signal
Transmembrane
Transmembrane helix
Vision
Feature
chain Peptidyl-prolyl cis-trans isomerase
Uniprot
A0A194RCB0
A0A2H1WSB3
A0A194PRM8
A0A0L7LST9
A0A212ESX9
A0A232FL48
+ More
A0A1W4X0L7 A0A1L8DTW1 Q16VU6 A0A182V9Q1 Q7Q137 A0A182HR92 A0A182KZ43 A0A182XGY3 A0A182Y6A1 A0A182NP22 A0A1B0GLX0 A0A182RWQ9 A0A182W8R3 A0A087ZN42 A0A2A3EHQ6 A0A182Q1Y4 A0A084VSR9 A0A182J2S2 E2C9W8 V9IJJ4 A0A158ND00 A0A0L7R6E0 A0A1J1I1K6 A0A182K951 A0A1W7R7Y6 A0A182P7Z6 W5J602 T1DRZ2 A0A182M1R3 A0A182SF46 A0A336KUL6 A0A0M8ZSS3 A0A1Q3G5B3 A0A139WGQ8 A0A139WHB1 A0A195D0J8 E0VPS3 W8B9L2 G9C5E4 A0A1B6M828 A0A026WGV9 A0A0K8UM76 A0A0K8U0Z0 A0A1B6MRG3 B3MM51 A0A0A9XF44 A0A146LVT8 T1GQ69 A0A1I8MY03 B4P2Z0 B3N8A1 A0A034VWL3 Q540W6 P15425 A0A0J9QUC6 Q29NN3 A0A1A9VXN4 B4GQP2 A0A1W4V2D3 B4ID30 A0A1B0B4N9 B4K1T6 B4N0P5 A0A1A9XDQ9 A0A1A9ZIX0 A0A182FWX4 A0A3B0JUF4 A0A1A9W0K5 B4JDU7 A0A1I8P8X2 A0A0N8EB86 A0A0P5L1J7 A0A0P5PMN4 A0A224XY85 B4KF64 A0A0P5JIN6 A0A0P5PW62 A0A0P5PMS8 A0A0P5XHJ4 A0A164S936 T1H9G1 P28517 A0A0N8C9T7 A0A0P5LI06 A0A0P5GMW9 A0A0P6BH72 A0A0P5LP10 A0A0P5KTI5 A0A0P5KY89 A0A0L0BQW7 A0A2S2NZR4 A0A2H8TMM6 A0A0P5AP13
A0A1W4X0L7 A0A1L8DTW1 Q16VU6 A0A182V9Q1 Q7Q137 A0A182HR92 A0A182KZ43 A0A182XGY3 A0A182Y6A1 A0A182NP22 A0A1B0GLX0 A0A182RWQ9 A0A182W8R3 A0A087ZN42 A0A2A3EHQ6 A0A182Q1Y4 A0A084VSR9 A0A182J2S2 E2C9W8 V9IJJ4 A0A158ND00 A0A0L7R6E0 A0A1J1I1K6 A0A182K951 A0A1W7R7Y6 A0A182P7Z6 W5J602 T1DRZ2 A0A182M1R3 A0A182SF46 A0A336KUL6 A0A0M8ZSS3 A0A1Q3G5B3 A0A139WGQ8 A0A139WHB1 A0A195D0J8 E0VPS3 W8B9L2 G9C5E4 A0A1B6M828 A0A026WGV9 A0A0K8UM76 A0A0K8U0Z0 A0A1B6MRG3 B3MM51 A0A0A9XF44 A0A146LVT8 T1GQ69 A0A1I8MY03 B4P2Z0 B3N8A1 A0A034VWL3 Q540W6 P15425 A0A0J9QUC6 Q29NN3 A0A1A9VXN4 B4GQP2 A0A1W4V2D3 B4ID30 A0A1B0B4N9 B4K1T6 B4N0P5 A0A1A9XDQ9 A0A1A9ZIX0 A0A182FWX4 A0A3B0JUF4 A0A1A9W0K5 B4JDU7 A0A1I8P8X2 A0A0N8EB86 A0A0P5L1J7 A0A0P5PMN4 A0A224XY85 B4KF64 A0A0P5JIN6 A0A0P5PW62 A0A0P5PMS8 A0A0P5XHJ4 A0A164S936 T1H9G1 P28517 A0A0N8C9T7 A0A0P5LI06 A0A0P5GMW9 A0A0P6BH72 A0A0P5LP10 A0A0P5KTI5 A0A0P5KY89 A0A0L0BQW7 A0A2S2NZR4 A0A2H8TMM6 A0A0P5AP13
EC Number
5.2.1.8
Pubmed
26354079
26227816
22118469
28648823
17510324
12364791
+ More
14747013 17210077 20966253 25244985 24438588 20798317 21347285 20920257 23761445 18362917 19820115 20566863 24495485 21835214 24508170 17994087 25401762 26823975 25315136 17550304 25348373 2493138 2664782 10731132 12537572 1707759 1644830 22936249 15632085 26108605
14747013 17210077 20966253 25244985 24438588 20798317 21347285 20920257 23761445 18362917 19820115 20566863 24495485 21835214 24508170 17994087 25401762 26823975 25315136 17550304 25348373 2493138 2664782 10731132 12537572 1707759 1644830 22936249 15632085 26108605
EMBL
KQ460398
KPJ15104.1
ODYU01010684
SOQ55960.1
KQ459601
KPI93790.1
+ More
JTDY01000158 KOB78538.1 AGBW02012699 OWR44598.1 NNAY01000063 OXU31392.1 GFDF01004203 JAV09881.1 CH477580 EAT38713.1 AAAB01008980 EAA13914.4 APCN01001659 AJVK01009166 KZ288242 PBC31238.1 AXCN02001964 ATLV01016140 KE525057 KFB41013.1 GL453904 EFN75266.1 JR048987 AEY60827.1 ADTU01012128 KQ414646 KOC66384.1 CVRI01000037 CRK93634.1 GEHC01000399 JAV47246.1 ADMH02002089 ETN59396.1 GAMD01000420 JAB01171.1 AXCM01000709 UFQS01000960 UFQT01000960 SSX07976.1 SSX28210.1 KQ435896 KOX69236.1 GFDL01000058 JAV34987.1 KQ971343 KYB27178.1 KYB27177.1 KQ977012 KYN06381.1 DS235371 EEB15379.1 GAMC01016689 JAB89866.1 HQ851385 AEV89763.1 GEBQ01007888 JAT32089.1 KK107213 EZA55302.1 GDHF01024711 JAI27603.1 GDHF01031947 JAI20367.1 GEBQ01001502 JAT38475.1 CH902620 EDV30866.1 GBHO01026172 GBHO01026171 JAG17432.1 JAG17433.1 GDHC01006808 JAQ11821.1 CAQQ02394226 CM000157 EDW87197.1 CH954177 EDV57288.1 GAKP01012737 GAKP01012735 JAC46215.1 AY119494 AAM50148.1 X14769 M22851 AE014134 CM002910 KMY87399.1 CH379059 EAL34185.2 CH479187 EDW39914.1 CH480829 EDW45456.1 JXJN01008377 CH917841 EDW04846.1 CH963920 EDW77658.1 OUUW01000004 SPP79110.1 CH916368 EDW03467.1 GDIQ01043780 JAN50957.1 GDIQ01177528 JAK74197.1 GDIQ01128253 JAL23473.1 GFTR01003377 JAW13049.1 CH933807 EDW13047.1 GDIQ01197824 JAK53901.1 GDIQ01129504 JAL22222.1 GDIQ01126806 JAL24920.1 GDIP01073820 JAM29895.1 LRGB01002076 KZS09378.1 ACPB03001101 GDIQ01100763 JAL50963.1 GDIQ01177529 JAK74196.1 GDIQ01242108 JAK09617.1 GDIP01014638 JAM89077.1 GDIQ01177526 JAK74199.1 GDIQ01180429 JAK71296.1 GDIQ01177527 JAK74198.1 JRES01001507 KNC22396.1 GGMR01009933 MBY22552.1 GFXV01003107 MBW14912.1 GDIP01210300 JAJ13102.1
JTDY01000158 KOB78538.1 AGBW02012699 OWR44598.1 NNAY01000063 OXU31392.1 GFDF01004203 JAV09881.1 CH477580 EAT38713.1 AAAB01008980 EAA13914.4 APCN01001659 AJVK01009166 KZ288242 PBC31238.1 AXCN02001964 ATLV01016140 KE525057 KFB41013.1 GL453904 EFN75266.1 JR048987 AEY60827.1 ADTU01012128 KQ414646 KOC66384.1 CVRI01000037 CRK93634.1 GEHC01000399 JAV47246.1 ADMH02002089 ETN59396.1 GAMD01000420 JAB01171.1 AXCM01000709 UFQS01000960 UFQT01000960 SSX07976.1 SSX28210.1 KQ435896 KOX69236.1 GFDL01000058 JAV34987.1 KQ971343 KYB27178.1 KYB27177.1 KQ977012 KYN06381.1 DS235371 EEB15379.1 GAMC01016689 JAB89866.1 HQ851385 AEV89763.1 GEBQ01007888 JAT32089.1 KK107213 EZA55302.1 GDHF01024711 JAI27603.1 GDHF01031947 JAI20367.1 GEBQ01001502 JAT38475.1 CH902620 EDV30866.1 GBHO01026172 GBHO01026171 JAG17432.1 JAG17433.1 GDHC01006808 JAQ11821.1 CAQQ02394226 CM000157 EDW87197.1 CH954177 EDV57288.1 GAKP01012737 GAKP01012735 JAC46215.1 AY119494 AAM50148.1 X14769 M22851 AE014134 CM002910 KMY87399.1 CH379059 EAL34185.2 CH479187 EDW39914.1 CH480829 EDW45456.1 JXJN01008377 CH917841 EDW04846.1 CH963920 EDW77658.1 OUUW01000004 SPP79110.1 CH916368 EDW03467.1 GDIQ01043780 JAN50957.1 GDIQ01177528 JAK74197.1 GDIQ01128253 JAL23473.1 GFTR01003377 JAW13049.1 CH933807 EDW13047.1 GDIQ01197824 JAK53901.1 GDIQ01129504 JAL22222.1 GDIQ01126806 JAL24920.1 GDIP01073820 JAM29895.1 LRGB01002076 KZS09378.1 ACPB03001101 GDIQ01100763 JAL50963.1 GDIQ01177529 JAK74196.1 GDIQ01242108 JAK09617.1 GDIP01014638 JAM89077.1 GDIQ01177526 JAK74199.1 GDIQ01180429 JAK71296.1 GDIQ01177527 JAK74198.1 JRES01001507 KNC22396.1 GGMR01009933 MBY22552.1 GFXV01003107 MBW14912.1 GDIP01210300 JAJ13102.1
Proteomes
UP000053240
UP000053268
UP000037510
UP000007151
UP000215335
UP000192223
+ More
UP000008820 UP000075903 UP000007062 UP000075840 UP000075882 UP000076407 UP000076408 UP000075884 UP000092462 UP000075900 UP000075920 UP000005203 UP000242457 UP000075886 UP000030765 UP000075880 UP000008237 UP000005205 UP000053825 UP000183832 UP000075881 UP000075885 UP000000673 UP000075883 UP000075901 UP000053105 UP000007266 UP000078542 UP000009046 UP000053097 UP000007801 UP000015102 UP000095301 UP000002282 UP000008711 UP000000803 UP000001819 UP000078200 UP000008744 UP000192221 UP000001292 UP000092460 UP000001070 UP000007798 UP000092443 UP000092445 UP000069272 UP000268350 UP000091820 UP000095300 UP000009192 UP000076858 UP000015103 UP000037069
UP000008820 UP000075903 UP000007062 UP000075840 UP000075882 UP000076407 UP000076408 UP000075884 UP000092462 UP000075900 UP000075920 UP000005203 UP000242457 UP000075886 UP000030765 UP000075880 UP000008237 UP000005205 UP000053825 UP000183832 UP000075881 UP000075885 UP000000673 UP000075883 UP000075901 UP000053105 UP000007266 UP000078542 UP000009046 UP000053097 UP000007801 UP000015102 UP000095301 UP000002282 UP000008711 UP000000803 UP000001819 UP000078200 UP000008744 UP000192221 UP000001292 UP000092460 UP000001070 UP000007798 UP000092443 UP000092445 UP000069272 UP000268350 UP000091820 UP000095300 UP000009192 UP000076858 UP000015103 UP000037069
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194RCB0
A0A2H1WSB3
A0A194PRM8
A0A0L7LST9
A0A212ESX9
A0A232FL48
+ More
A0A1W4X0L7 A0A1L8DTW1 Q16VU6 A0A182V9Q1 Q7Q137 A0A182HR92 A0A182KZ43 A0A182XGY3 A0A182Y6A1 A0A182NP22 A0A1B0GLX0 A0A182RWQ9 A0A182W8R3 A0A087ZN42 A0A2A3EHQ6 A0A182Q1Y4 A0A084VSR9 A0A182J2S2 E2C9W8 V9IJJ4 A0A158ND00 A0A0L7R6E0 A0A1J1I1K6 A0A182K951 A0A1W7R7Y6 A0A182P7Z6 W5J602 T1DRZ2 A0A182M1R3 A0A182SF46 A0A336KUL6 A0A0M8ZSS3 A0A1Q3G5B3 A0A139WGQ8 A0A139WHB1 A0A195D0J8 E0VPS3 W8B9L2 G9C5E4 A0A1B6M828 A0A026WGV9 A0A0K8UM76 A0A0K8U0Z0 A0A1B6MRG3 B3MM51 A0A0A9XF44 A0A146LVT8 T1GQ69 A0A1I8MY03 B4P2Z0 B3N8A1 A0A034VWL3 Q540W6 P15425 A0A0J9QUC6 Q29NN3 A0A1A9VXN4 B4GQP2 A0A1W4V2D3 B4ID30 A0A1B0B4N9 B4K1T6 B4N0P5 A0A1A9XDQ9 A0A1A9ZIX0 A0A182FWX4 A0A3B0JUF4 A0A1A9W0K5 B4JDU7 A0A1I8P8X2 A0A0N8EB86 A0A0P5L1J7 A0A0P5PMN4 A0A224XY85 B4KF64 A0A0P5JIN6 A0A0P5PW62 A0A0P5PMS8 A0A0P5XHJ4 A0A164S936 T1H9G1 P28517 A0A0N8C9T7 A0A0P5LI06 A0A0P5GMW9 A0A0P6BH72 A0A0P5LP10 A0A0P5KTI5 A0A0P5KY89 A0A0L0BQW7 A0A2S2NZR4 A0A2H8TMM6 A0A0P5AP13
A0A1W4X0L7 A0A1L8DTW1 Q16VU6 A0A182V9Q1 Q7Q137 A0A182HR92 A0A182KZ43 A0A182XGY3 A0A182Y6A1 A0A182NP22 A0A1B0GLX0 A0A182RWQ9 A0A182W8R3 A0A087ZN42 A0A2A3EHQ6 A0A182Q1Y4 A0A084VSR9 A0A182J2S2 E2C9W8 V9IJJ4 A0A158ND00 A0A0L7R6E0 A0A1J1I1K6 A0A182K951 A0A1W7R7Y6 A0A182P7Z6 W5J602 T1DRZ2 A0A182M1R3 A0A182SF46 A0A336KUL6 A0A0M8ZSS3 A0A1Q3G5B3 A0A139WGQ8 A0A139WHB1 A0A195D0J8 E0VPS3 W8B9L2 G9C5E4 A0A1B6M828 A0A026WGV9 A0A0K8UM76 A0A0K8U0Z0 A0A1B6MRG3 B3MM51 A0A0A9XF44 A0A146LVT8 T1GQ69 A0A1I8MY03 B4P2Z0 B3N8A1 A0A034VWL3 Q540W6 P15425 A0A0J9QUC6 Q29NN3 A0A1A9VXN4 B4GQP2 A0A1W4V2D3 B4ID30 A0A1B0B4N9 B4K1T6 B4N0P5 A0A1A9XDQ9 A0A1A9ZIX0 A0A182FWX4 A0A3B0JUF4 A0A1A9W0K5 B4JDU7 A0A1I8P8X2 A0A0N8EB86 A0A0P5L1J7 A0A0P5PMN4 A0A224XY85 B4KF64 A0A0P5JIN6 A0A0P5PW62 A0A0P5PMS8 A0A0P5XHJ4 A0A164S936 T1H9G1 P28517 A0A0N8C9T7 A0A0P5LI06 A0A0P5GMW9 A0A0P6BH72 A0A0P5LP10 A0A0P5KTI5 A0A0P5KY89 A0A0L0BQW7 A0A2S2NZR4 A0A2H8TMM6 A0A0P5AP13
PDB
4FRV
E-value=6.80917e-38,
Score=392
Ontologies
GO
Topology
Subcellular location
Membrane
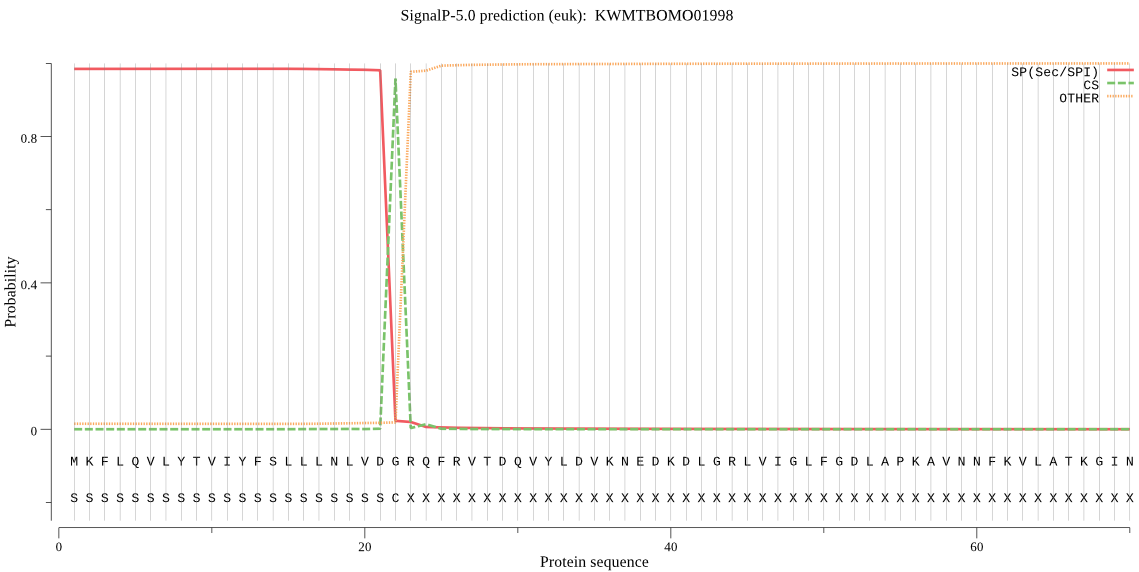
SignalP
Position: 1 - 22,
Likelihood: 0.984635
Length:
236
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.18755
Exp number, first 60 AAs:
5.16501
Total prob of N-in:
0.13427
outside
1 - 206
TMhelix
207 - 229
inside
230 - 236

Population Genetic Test Statistics
Pi
343.620443
Theta
168.673644
Tajima's D
3.258319
CLR
0.374615
CSRT
0.987900604969752
Interpretation
Uncertain
