Gene
KWMTBOMO01992
Pre Gene Modal
BGIBMGA013559
Annotation
PREDICTED:_pyruvate_kinase-like_[Bombyx_mori]
Full name
Pyruvate kinase
Location in the cell
Cytoplasmic Reliability : 2.703
Sequence
CDS
ATGTCACTACCTTGGCATATTCCATTAGAGGCTGCACCCTGTTTTAAAGAAAAACTCAATGCGGTACAACCAACATTGCTCAAACATTACTGTAACATTAATGTACACGAAAATCCTTGCGAGGAGCTACAAACGGGACTCATGTGTGACATAGGACTTGATAACAGAGAACCAGCTACAATACAAAGACTAATAGCTTCAGGGATGACTGTAGCACGATTAAATATGCAAAATTTAGAACCAGAAGCATGCGCTCAACTTATTCAAAGCATCCGACAAGCTGTGTATAATTATAGTTTAGAGCTAGATTTTGTTTATCCTCTAGCTGTCGTGGTTGATGTCAAAGGTCCAGATATTGTCACAGGAGGACTGAAAAATGGTCCACATGCTACTGTTGAGCTTTTACAAACTGAAACAATTCGGCTAACAATAGATCCCAGTTGGAGGGACTGTGGGACATCGGAATGCCTTTACGTAGGATATGATCAATTGACAGATTTGAATACAGATGATATAATTTATATAGATAGCTTATCACCAGGAAAAATCAAACTAGTTGTTACAGAAGTGGGAGATGATTCCTTAGAATGTCTTATTGTTATTGGCGGTACCATTGGAGCTAGTATGGCAGTGCGTATATCATCAATACCTATGGAAGTACAAAGCTGTAGACGAGATGATAGTGCTGATTCCATTTCCGATATTATTAGTTCAAGCCAAGCATTTGAACATTTAGATGAACAAGTTAATTGGGCTACAGCTTCAGATGTTGATGCTTTATTAGTACCCAACTGCCAAGGATCACATGATATCAAACAAATTAAAGAAATGCTTTCAGAGAAAGGAAAACACATTCTTGTCCTTGCTTGTATAGATACAGTCCTAGGGCTTGATAATATAGATGAAGTACTGGAAGAAGCAGATGGCATTTATGTTGATCGATCTGTTTTGAGTACTGATTTACCAGTAGAAAAAATATTTTTGGCCCAAAAACAGATTCTTGCGAAGTGTAATGCAGTGGGAAAACCTTGCTTATGCAAAGCAGTACTTAATGAACAAATACCAACACTATGCGTAACAGATTTAGCAAATCTCGTTTTAGACGGAGCAGATGTTCTTTCTTTAGAACTTCATTATGATTCACCCCTAAAGAAATTTGCACCATTGTATGACGCTATTCGTATGGCCGAACATTGCTTAGGAGCAGCTGGAGTAATTTGCAGACAAGCAGAAAGAATAACATGGAAACCTAATATTCATGGAAATCAAGATTTACTTAAAAATCGTAACGAGGATCCAACAAAATCTGTTTGTATTACAGCAGCTGAATTGGCAGCATTGTCGCAAGCAGTTGTAATTATATGCTTAACGAATTCAGGTCGTACAGCTAAAATTTTATCCTATATAAAACCGGTGTGTCCTATCGTTGCCGTGACTAGAACTTGTCATACAGCTAGGCAATTGCGATTTTGGCGTGGCGTACGCGCTGTACATTATTTTGAATCTCCTAAAGGAAATTGGTTAAATGAAGTAGAATCCAGGATATTTGCTGCATTAAACTACTGTAAAGCAAAAAGAATGCTACGTGCAGGTGATGCATACGTTATCATTTCTGGATCACGTCGTGGTGTAGGATATTGTGATTCGGTCAAACTTTTATTTGCAAGTGTTCGGGAAGCAACCTATTTGGAATAG
Protein
MSLPWHIPLEAAPCFKEKLNAVQPTLLKHYCNINVHENPCEELQTGLMCDIGLDNREPATIQRLIASGMTVARLNMQNLEPEACAQLIQSIRQAVYNYSLELDFVYPLAVVVDVKGPDIVTGGLKNGPHATVELLQTETIRLTIDPSWRDCGTSECLYVGYDQLTDLNTDDIIYIDSLSPGKIKLVVTEVGDDSLECLIVIGGTIGASMAVRISSIPMEVQSCRRDDSADSISDIISSSQAFEHLDEQVNWATASDVDALLVPNCQGSHDIKQIKEMLSEKGKHILVLACIDTVLGLDNIDEVLEEADGIYVDRSVLSTDLPVEKIFLAQKQILAKCNAVGKPCLCKAVLNEQIPTLCVTDLANLVLDGADVLSLELHYDSPLKKFAPLYDAIRMAEHCLGAAGVICRQAERITWKPNIHGNQDLLKNRNEDPTKSVCITAAELAALSQAVVIICLTNSGRTAKILSYIKPVCPIVAVTRTCHTARQLRFWRGVRAVHYFESPKGNWLNEVESRIFAALNYCKAKRMLRAGDAYVIISGSRRGVGYCDSVKLLFASVREATYLE
Summary
Catalytic Activity
ATP + pyruvate = ADP + H(+) + phosphoenolpyruvate
Similarity
Belongs to the pyruvate kinase family.
Uniprot
H9JVJ6
A0A212ESY9
A0A194RCC0
A0A0L7L3J4
A0A2A4J6V0
A0A194PLC6
+ More
A0A2H1VS12 A0A0P5T0A6 A0A0P5BE35 A0A0N8DAG1 A0A0P5QRU6 A0A0P5IXU7 J3JXD8 A0A0P4ZYY1 A0A0P4Z8B1 A0A0N8A026 A0A0P5WDT8 A0A2Z5REH9 A0A0P5PF75 A0A0P5EN83 A0A162DCG0 A0A0P5BYV3 A0A0P5TN41 A0A0P4ZTD6 A0A0P5R9S5 A0A0N8D3D1 A0A0P6H8M0 A0A0P5G5H3 A0A2Z5REH8 A0A0P5I6S5 A0A0P5CUB7 A0A0P5RR23 A0A0P5ETT1 A0A0P5PVF5 A0A0P5A0V7 A0A0P5JI55 A0A0P4YPL7 A0A0P5Q9B3 A0A0P5AW78 A0A0P5HLS1 A0A0P5CR60 A0A0P6GNB8 A0A0P4Z602 A0A0P5X7T5 A0A0P5THW1 A0A0P5BM20 A0A0P5WLB5 A0A0N8E2I2 U5ESC2 E9HUQ2 A0A0P6ABY5 A0A0P4WPS4 A0A232ESM8 A0A139WAK3 N6T4X5 A0A182NN15 V5SJ07 A0A1Y1L4F9 K7J236 A0A182VFI7 A0A182LNM2 D6X528 A0A139WAJ1 Q7PPE7 A0A1Y9GJK0 A0A1S4GLJ9 A0A182XDN4 A0A182IDE2 A0A182UE55 A0A182MNQ6 A0A0A9XHK7 A0A1W5YLK4 A0A1X9QDT2 A0A182VPN6 A0A0A9YKR8 V5I8G0 A0A2J7RLW1 A0A182SYI1 A0A0C5QRX0 A0A0A9YVU7 A0A2J7RLY6 A0A1Y9GJY2 A0A182RED7 A0A0K8TLX8 A0A226EGV1 A0A2J7RLV9 A0A084W7S0 A0A2H8TDH4 A0A182PWA9 A0A182JJQ1 J9JR33 A0A2J7RLW3 A0A182KAF7 A0A2I9LPH0 I5ANV2 A0A0R3NNH8 B5DYU6 A0A2L2XYS9 I4DJQ4 A0A1Y1LFI1
A0A2H1VS12 A0A0P5T0A6 A0A0P5BE35 A0A0N8DAG1 A0A0P5QRU6 A0A0P5IXU7 J3JXD8 A0A0P4ZYY1 A0A0P4Z8B1 A0A0N8A026 A0A0P5WDT8 A0A2Z5REH9 A0A0P5PF75 A0A0P5EN83 A0A162DCG0 A0A0P5BYV3 A0A0P5TN41 A0A0P4ZTD6 A0A0P5R9S5 A0A0N8D3D1 A0A0P6H8M0 A0A0P5G5H3 A0A2Z5REH8 A0A0P5I6S5 A0A0P5CUB7 A0A0P5RR23 A0A0P5ETT1 A0A0P5PVF5 A0A0P5A0V7 A0A0P5JI55 A0A0P4YPL7 A0A0P5Q9B3 A0A0P5AW78 A0A0P5HLS1 A0A0P5CR60 A0A0P6GNB8 A0A0P4Z602 A0A0P5X7T5 A0A0P5THW1 A0A0P5BM20 A0A0P5WLB5 A0A0N8E2I2 U5ESC2 E9HUQ2 A0A0P6ABY5 A0A0P4WPS4 A0A232ESM8 A0A139WAK3 N6T4X5 A0A182NN15 V5SJ07 A0A1Y1L4F9 K7J236 A0A182VFI7 A0A182LNM2 D6X528 A0A139WAJ1 Q7PPE7 A0A1Y9GJK0 A0A1S4GLJ9 A0A182XDN4 A0A182IDE2 A0A182UE55 A0A182MNQ6 A0A0A9XHK7 A0A1W5YLK4 A0A1X9QDT2 A0A182VPN6 A0A0A9YKR8 V5I8G0 A0A2J7RLW1 A0A182SYI1 A0A0C5QRX0 A0A0A9YVU7 A0A2J7RLY6 A0A1Y9GJY2 A0A182RED7 A0A0K8TLX8 A0A226EGV1 A0A2J7RLV9 A0A084W7S0 A0A2H8TDH4 A0A182PWA9 A0A182JJQ1 J9JR33 A0A2J7RLW3 A0A182KAF7 A0A2I9LPH0 I5ANV2 A0A0R3NNH8 B5DYU6 A0A2L2XYS9 I4DJQ4 A0A1Y1LFI1
EC Number
2.7.1.40
Pubmed
EMBL
BABH01038975
BABH01038976
AGBW02012699
OWR44589.1
KQ460398
KPJ15114.1
+ More
JTDY01003243 KOB69891.1 NWSH01002987 PCG67143.1 KQ459601 KPI93798.1 ODYU01004097 SOQ43609.1 GDIP01132793 JAL70921.1 GDIP01185840 JAJ37562.1 GDIP01052959 JAM50756.1 GDIQ01130741 JAL20985.1 GDIQ01207910 JAK43815.1 APGK01052723 APGK01052724 BT127907 KB741213 KB631753 AEE62869.1 ENN72684.1 ERL85807.1 GDIP01219278 JAJ04124.1 GDIP01228677 JAI94724.1 GDIP01195729 JAJ27673.1 GDIP01088202 JAM15513.1 FX983728 BAX07227.1 GDIQ01129439 JAL22287.1 GDIP01145591 JAJ77811.1 LRGB01002864 KZS05735.1 GDIP01182684 JAJ40718.1 GDIP01127394 JAL76320.1 GDIP01221792 JAJ01610.1 GDIQ01103836 JAL47890.1 GDIP01072799 JAM30916.1 GDIQ01023110 JAN71627.1 GDIQ01246938 JAK04787.1 FX983727 BAX07226.1 GDIQ01217946 JAK33779.1 GDIP01165586 JAJ57816.1 GDIQ01115884 JAL35842.1 GDIQ01270260 JAJ81464.1 GDIQ01129438 JAL22288.1 GDIP01209933 JAJ13469.1 GDIQ01198898 JAK52827.1 GDIP01228676 GDIP01201525 GDIP01130092 GDIP01095448 JAI94725.1 GDIQ01126637 JAL25089.1 GDIP01197791 JAJ25611.1 GDIQ01225553 JAK26172.1 GDIP01167933 JAJ55469.1 GDIQ01032763 JAN61974.1 GDIP01221791 JAJ01611.1 GDIP01088201 JAM15514.1 GDIP01126000 JAL77714.1 GDIP01182683 JAJ40719.1 GDIP01084539 JAM19176.1 GDIQ01068212 JAN26525.1 GANO01002423 JAB57448.1 GL732818 EFX64530.1 GDIP01032046 JAM71669.1 GDIP01253375 JAI70026.1 NNAY01002419 OXU21317.1 KQ971382 KYB24935.1 ENN72683.1 KF647624 AHB50486.1 GEZM01068639 JAV66845.1 EEZ97193.2 KYB24934.1 AAAB01008952 EAA10555.6 APCN01005967 AXCM01000245 GBHO01024498 GBHO01023780 GBHO01010803 GBHO01010230 GBHO01006495 GBHO01006493 JAG19106.1 JAG19824.1 JAG32801.1 JAG33374.1 JAG37109.1 JAG37111.1 KX147661 ARI45065.1 KY412773 ARQ20739.1 GBHO01010825 GBHO01010824 GBHO01010805 GBHO01010307 GBHO01010289 GBHO01009918 GDHC01007241 JAG32779.1 JAG32780.1 JAG32799.1 JAG33297.1 JAG33315.1 JAG33686.1 JAQ11388.1 GALX01004750 JAB63716.1 NEVH01002616 PNF41818.1 KM609491 AJQ30115.1 GBHO01023781 GBHO01023776 GBHO01023775 GBHO01010806 GBHO01009950 GBHO01006502 GBHO01006497 JAG19823.1 JAG19828.1 JAG19829.1 JAG32798.1 JAG33654.1 JAG37102.1 JAG37107.1 PNF41819.1 GDAI01002239 JAI15364.1 LNIX01000004 OXA56448.1 PNF41817.1 ATLV01021293 KE525316 KFB46264.1 GFXV01000361 MBW12166.1 ABLF02017366 PNF41820.1 GFWZ01000284 MBW20274.1 CM000070 EIM52637.2 KRT00326.1 EDY68021.1 IAAA01002410 LAA01038.1 AK401522 BAM18144.1 GEZM01057153 JAV72422.1
JTDY01003243 KOB69891.1 NWSH01002987 PCG67143.1 KQ459601 KPI93798.1 ODYU01004097 SOQ43609.1 GDIP01132793 JAL70921.1 GDIP01185840 JAJ37562.1 GDIP01052959 JAM50756.1 GDIQ01130741 JAL20985.1 GDIQ01207910 JAK43815.1 APGK01052723 APGK01052724 BT127907 KB741213 KB631753 AEE62869.1 ENN72684.1 ERL85807.1 GDIP01219278 JAJ04124.1 GDIP01228677 JAI94724.1 GDIP01195729 JAJ27673.1 GDIP01088202 JAM15513.1 FX983728 BAX07227.1 GDIQ01129439 JAL22287.1 GDIP01145591 JAJ77811.1 LRGB01002864 KZS05735.1 GDIP01182684 JAJ40718.1 GDIP01127394 JAL76320.1 GDIP01221792 JAJ01610.1 GDIQ01103836 JAL47890.1 GDIP01072799 JAM30916.1 GDIQ01023110 JAN71627.1 GDIQ01246938 JAK04787.1 FX983727 BAX07226.1 GDIQ01217946 JAK33779.1 GDIP01165586 JAJ57816.1 GDIQ01115884 JAL35842.1 GDIQ01270260 JAJ81464.1 GDIQ01129438 JAL22288.1 GDIP01209933 JAJ13469.1 GDIQ01198898 JAK52827.1 GDIP01228676 GDIP01201525 GDIP01130092 GDIP01095448 JAI94725.1 GDIQ01126637 JAL25089.1 GDIP01197791 JAJ25611.1 GDIQ01225553 JAK26172.1 GDIP01167933 JAJ55469.1 GDIQ01032763 JAN61974.1 GDIP01221791 JAJ01611.1 GDIP01088201 JAM15514.1 GDIP01126000 JAL77714.1 GDIP01182683 JAJ40719.1 GDIP01084539 JAM19176.1 GDIQ01068212 JAN26525.1 GANO01002423 JAB57448.1 GL732818 EFX64530.1 GDIP01032046 JAM71669.1 GDIP01253375 JAI70026.1 NNAY01002419 OXU21317.1 KQ971382 KYB24935.1 ENN72683.1 KF647624 AHB50486.1 GEZM01068639 JAV66845.1 EEZ97193.2 KYB24934.1 AAAB01008952 EAA10555.6 APCN01005967 AXCM01000245 GBHO01024498 GBHO01023780 GBHO01010803 GBHO01010230 GBHO01006495 GBHO01006493 JAG19106.1 JAG19824.1 JAG32801.1 JAG33374.1 JAG37109.1 JAG37111.1 KX147661 ARI45065.1 KY412773 ARQ20739.1 GBHO01010825 GBHO01010824 GBHO01010805 GBHO01010307 GBHO01010289 GBHO01009918 GDHC01007241 JAG32779.1 JAG32780.1 JAG32799.1 JAG33297.1 JAG33315.1 JAG33686.1 JAQ11388.1 GALX01004750 JAB63716.1 NEVH01002616 PNF41818.1 KM609491 AJQ30115.1 GBHO01023781 GBHO01023776 GBHO01023775 GBHO01010806 GBHO01009950 GBHO01006502 GBHO01006497 JAG19823.1 JAG19828.1 JAG19829.1 JAG32798.1 JAG33654.1 JAG37102.1 JAG37107.1 PNF41819.1 GDAI01002239 JAI15364.1 LNIX01000004 OXA56448.1 PNF41817.1 ATLV01021293 KE525316 KFB46264.1 GFXV01000361 MBW12166.1 ABLF02017366 PNF41820.1 GFWZ01000284 MBW20274.1 CM000070 EIM52637.2 KRT00326.1 EDY68021.1 IAAA01002410 LAA01038.1 AK401522 BAM18144.1 GEZM01057153 JAV72422.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000037510
UP000218220
UP000053268
+ More
UP000019118 UP000030742 UP000076858 UP000000305 UP000215335 UP000007266 UP000075884 UP000002358 UP000075903 UP000075882 UP000007062 UP000075840 UP000076407 UP000075902 UP000075883 UP000075920 UP000235965 UP000075901 UP000075900 UP000198287 UP000030765 UP000075885 UP000075880 UP000007819 UP000075881 UP000001819
UP000019118 UP000030742 UP000076858 UP000000305 UP000215335 UP000007266 UP000075884 UP000002358 UP000075903 UP000075882 UP000007062 UP000075840 UP000076407 UP000075902 UP000075883 UP000075920 UP000235965 UP000075901 UP000075900 UP000198287 UP000030765 UP000075885 UP000075880 UP000007819 UP000075881 UP000001819
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JVJ6
A0A212ESY9
A0A194RCC0
A0A0L7L3J4
A0A2A4J6V0
A0A194PLC6
+ More
A0A2H1VS12 A0A0P5T0A6 A0A0P5BE35 A0A0N8DAG1 A0A0P5QRU6 A0A0P5IXU7 J3JXD8 A0A0P4ZYY1 A0A0P4Z8B1 A0A0N8A026 A0A0P5WDT8 A0A2Z5REH9 A0A0P5PF75 A0A0P5EN83 A0A162DCG0 A0A0P5BYV3 A0A0P5TN41 A0A0P4ZTD6 A0A0P5R9S5 A0A0N8D3D1 A0A0P6H8M0 A0A0P5G5H3 A0A2Z5REH8 A0A0P5I6S5 A0A0P5CUB7 A0A0P5RR23 A0A0P5ETT1 A0A0P5PVF5 A0A0P5A0V7 A0A0P5JI55 A0A0P4YPL7 A0A0P5Q9B3 A0A0P5AW78 A0A0P5HLS1 A0A0P5CR60 A0A0P6GNB8 A0A0P4Z602 A0A0P5X7T5 A0A0P5THW1 A0A0P5BM20 A0A0P5WLB5 A0A0N8E2I2 U5ESC2 E9HUQ2 A0A0P6ABY5 A0A0P4WPS4 A0A232ESM8 A0A139WAK3 N6T4X5 A0A182NN15 V5SJ07 A0A1Y1L4F9 K7J236 A0A182VFI7 A0A182LNM2 D6X528 A0A139WAJ1 Q7PPE7 A0A1Y9GJK0 A0A1S4GLJ9 A0A182XDN4 A0A182IDE2 A0A182UE55 A0A182MNQ6 A0A0A9XHK7 A0A1W5YLK4 A0A1X9QDT2 A0A182VPN6 A0A0A9YKR8 V5I8G0 A0A2J7RLW1 A0A182SYI1 A0A0C5QRX0 A0A0A9YVU7 A0A2J7RLY6 A0A1Y9GJY2 A0A182RED7 A0A0K8TLX8 A0A226EGV1 A0A2J7RLV9 A0A084W7S0 A0A2H8TDH4 A0A182PWA9 A0A182JJQ1 J9JR33 A0A2J7RLW3 A0A182KAF7 A0A2I9LPH0 I5ANV2 A0A0R3NNH8 B5DYU6 A0A2L2XYS9 I4DJQ4 A0A1Y1LFI1
A0A2H1VS12 A0A0P5T0A6 A0A0P5BE35 A0A0N8DAG1 A0A0P5QRU6 A0A0P5IXU7 J3JXD8 A0A0P4ZYY1 A0A0P4Z8B1 A0A0N8A026 A0A0P5WDT8 A0A2Z5REH9 A0A0P5PF75 A0A0P5EN83 A0A162DCG0 A0A0P5BYV3 A0A0P5TN41 A0A0P4ZTD6 A0A0P5R9S5 A0A0N8D3D1 A0A0P6H8M0 A0A0P5G5H3 A0A2Z5REH8 A0A0P5I6S5 A0A0P5CUB7 A0A0P5RR23 A0A0P5ETT1 A0A0P5PVF5 A0A0P5A0V7 A0A0P5JI55 A0A0P4YPL7 A0A0P5Q9B3 A0A0P5AW78 A0A0P5HLS1 A0A0P5CR60 A0A0P6GNB8 A0A0P4Z602 A0A0P5X7T5 A0A0P5THW1 A0A0P5BM20 A0A0P5WLB5 A0A0N8E2I2 U5ESC2 E9HUQ2 A0A0P6ABY5 A0A0P4WPS4 A0A232ESM8 A0A139WAK3 N6T4X5 A0A182NN15 V5SJ07 A0A1Y1L4F9 K7J236 A0A182VFI7 A0A182LNM2 D6X528 A0A139WAJ1 Q7PPE7 A0A1Y9GJK0 A0A1S4GLJ9 A0A182XDN4 A0A182IDE2 A0A182UE55 A0A182MNQ6 A0A0A9XHK7 A0A1W5YLK4 A0A1X9QDT2 A0A182VPN6 A0A0A9YKR8 V5I8G0 A0A2J7RLW1 A0A182SYI1 A0A0C5QRX0 A0A0A9YVU7 A0A2J7RLY6 A0A1Y9GJY2 A0A182RED7 A0A0K8TLX8 A0A226EGV1 A0A2J7RLV9 A0A084W7S0 A0A2H8TDH4 A0A182PWA9 A0A182JJQ1 J9JR33 A0A2J7RLW3 A0A182KAF7 A0A2I9LPH0 I5ANV2 A0A0R3NNH8 B5DYU6 A0A2L2XYS9 I4DJQ4 A0A1Y1LFI1
PDB
6DU6
E-value=4.24301e-70,
Score=674
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00230 Purine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00230 Purine metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
PANTHER
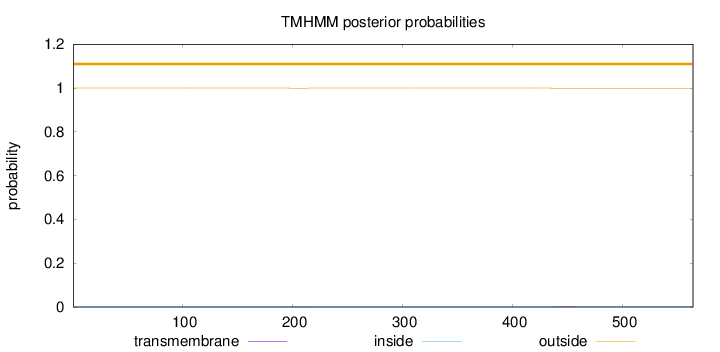
Topology
Length:
564
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0628300000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00029
outside
1 - 564
Population Genetic Test Statistics
Pi
281.696612
Theta
229.520112
Tajima's D
0.603087
CLR
0
CSRT
0.544922753862307
Interpretation
Uncertain
