Gene
KWMTBOMO01987
Annotation
PREDICTED:_mucin-2-like_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.525
Sequence
CDS
ATGCACATTCCAAAAGAGAAGCGCCTCAAATGGAATAAAAAGGCTGTAAAAGGGATTCTAGTTGGTTTCCCAGAGGATGTAAAAGGTTATCGAGTCTACAATCCAAGCACTAAGGATATCACAACAAGCCGTGATGTGATCATTATAGAAAATGCCAAACAAACAGAAAGTGTAGTCCAGATTGAGGAAACTGAACAATCCAGTGAGGATTCAGTGGGAGACACATCGTATGAATCAGGAAGTGAAATTTTTCATGATGATAATGATGAGACTTATGTACCAGATGATTCTTCTGATGACAGTGTGGAAAGTGTTGCTCTAAAAGGACCAGATAAGGAACAGTGGTTAGAGGCAATAAGGGACGAATTACAGTGTTTTCAAGATAATGATGCTTGGGAACTTACTGATGCACCTAAAGACAGTACTATAGTTAAATGTAGATGGGTTTTGTGTAAGAAATATGATAGTGAAAGTAAGATTAGGTTCCGAGCCAGACTTGTAGCTAAAGGTTTTTCCCAGAAACAAGGAGTAGATTATAAGGAAACCTTCTCGCCTGCTGTTAGACATACAACATTACGACTTTTGTTTTCACTATGA
Protein
MHIPKEKRLKWNKKAVKGILVGFPEDVKGYRVYNPSTKDITTSRDVIIIENAKQTESVVQIEETEQSSEDSVGDTSYESGSEIFHDDNDETYVPDDSSDDSVESVALKGPDKEQWLEAIRDELQCFQDNDAWELTDAPKDSTIVKCRWVLCKKYDSESKIRFRARLVAKGFSQKQGVDYKETFSPAVRHTTLRLLFSL
Summary
Similarity
Belongs to the terpene synthase family.
Uniprot
V5GAG2
A0A1Y1NAZ7
A0A1Y1N668
A0A1Y1KL90
X1X3E5
X1X2J4
+ More
J9L0N0 X1WZA6 A0A0J7K6W1 X1WT18 A0A0A9XV68 A0A0A9XY84 A0A146LH62 B8YLY3 A0A2N9H4R5 A0A151R6U8 A0A151UDA6 A0A085N0V2 B8YLY6 A0A2N9IR42 A0A0N1ICY1 B8YLY4 A0A151RL25 A0A2N9EK88 A0A2N9GG47 A0A2N9GZK0 A0A2N9EH79 A0A2N9HHJ9 A0A2N9I807 A0A2N9JAE7 A0A2N9H7C1 A0A2Z6N9Q0 A0A2N9HY48 A0A2N9FGL6 A0A2N9EYD8 A0A2N9ING1 A0A0V0U113 A0A2N9IGI9 A0A2K3NH55 A0A2N9IGN4 A0A2N9HJF5 A0A2N9FL83 A0A2N9FVE8 A0A2N9FRN5 A0A2N9HZR4 A0A151R2G6 A0A151RJN4 A0A151TKT1 A0A2N9FPD9 A0A151SDY6 A0A2N9EER7 A0A2N9EPH6 A0A0J7KH47 A0A2N9FZ01 A0A2N9HIB6 A0A2N9I1P0 A0A2N9I0K6 A0A2N9I9F7 A0A2N9IB18 A0A2N9F910 A0A2N9HQS0 A5AWP3 A0A2Z6N4D2 A0A2K3L574 A5B0R8 A0A2N9J0X3 A0A1Y3E9R1 A0A0J7K9V7 A0A0J7KE66 A0A151R8W9 A0A059QBK0 A0A1Y1JXD5 A0A1R3G990 A5B9M8 A5BMR0 X1XSG1 A0A2Z6N7E5 A5BR97 A0A2N9J5Y1 A0A0J7K724 K7JAV0 A0A2N9G211 A0A2N9GNZ7 A0A1R3GTE9 A0A3P1N2J7 A0A2N9HLW0 A0A151RQW4 A0A1U8MHG7 A0A151TAK4 A0A2Z6PCB3 A0A2Z6NRJ0 A0A2N9HJ44 A0A251VGU8 A0A1Y1LVN7 A0A1Y1MYP5
J9L0N0 X1WZA6 A0A0J7K6W1 X1WT18 A0A0A9XV68 A0A0A9XY84 A0A146LH62 B8YLY3 A0A2N9H4R5 A0A151R6U8 A0A151UDA6 A0A085N0V2 B8YLY6 A0A2N9IR42 A0A0N1ICY1 B8YLY4 A0A151RL25 A0A2N9EK88 A0A2N9GG47 A0A2N9GZK0 A0A2N9EH79 A0A2N9HHJ9 A0A2N9I807 A0A2N9JAE7 A0A2N9H7C1 A0A2Z6N9Q0 A0A2N9HY48 A0A2N9FGL6 A0A2N9EYD8 A0A2N9ING1 A0A0V0U113 A0A2N9IGI9 A0A2K3NH55 A0A2N9IGN4 A0A2N9HJF5 A0A2N9FL83 A0A2N9FVE8 A0A2N9FRN5 A0A2N9HZR4 A0A151R2G6 A0A151RJN4 A0A151TKT1 A0A2N9FPD9 A0A151SDY6 A0A2N9EER7 A0A2N9EPH6 A0A0J7KH47 A0A2N9FZ01 A0A2N9HIB6 A0A2N9I1P0 A0A2N9I0K6 A0A2N9I9F7 A0A2N9IB18 A0A2N9F910 A0A2N9HQS0 A5AWP3 A0A2Z6N4D2 A0A2K3L574 A5B0R8 A0A2N9J0X3 A0A1Y3E9R1 A0A0J7K9V7 A0A0J7KE66 A0A151R8W9 A0A059QBK0 A0A1Y1JXD5 A0A1R3G990 A5B9M8 A5BMR0 X1XSG1 A0A2Z6N7E5 A5BR97 A0A2N9J5Y1 A0A0J7K724 K7JAV0 A0A2N9G211 A0A2N9GNZ7 A0A1R3GTE9 A0A3P1N2J7 A0A2N9HLW0 A0A151RQW4 A0A1U8MHG7 A0A151TAK4 A0A2Z6PCB3 A0A2Z6NRJ0 A0A2N9HJ44 A0A251VGU8 A0A1Y1LVN7 A0A1Y1MYP5
Pubmed
EMBL
GALX01001351
JAB67115.1
GEZM01011855
JAV93396.1
GEZM01011856
JAV93393.1
+ More
GEZM01082909 JAV61231.1 ABLF02062902 ABLF02031072 ABLF02059092 ABLF02060653 ABLF02023757 ABLF02028466 ABLF02059696 ABLF02055865 LBMM01012645 KMQ86082.1 ABLF02014335 ABLF02060220 GBHO01019780 JAG23824.1 GBHO01019781 JAG23823.1 GDHC01011760 JAQ06869.1 FJ544851 ACL97383.1 OIVN01002812 SPD06621.1 KQ484038 KYP38095.1 AGCT01017703 KYP77285.1 KL367580 KFD63098.1 FJ544856 ACL97386.1 OIVN01006160 SPD26619.1 KQ460882 KPJ11482.1 FJ544853 ACL97384.1 KQ483680 KYP43165.1 OIVN01000147 SPC75223.1 OIVN01001850 SPC98251.1 OIVN01002946 SPD07686.1 OIVN01000089 SPC74041.1 OIVN01003423 SPD11099.1 OIVN01004957 SPD20159.1 OIVN01006462 SPD33594.1 OIVN01002925 SPD07540.1 DF973531 GAU33542.1 OIVN01004307 SPD16591.1 OIVN01000823 SPC86071.1 OIVN01000418 SPC79865.1 OIVN01006125 SPD25623.1 JYDJ01000099 KRX44323.1 OIVN01005591 SPD23220.1 ASHM01021274 PNY02378.1 OIVN01005657 SPD23465.1 OIVN01003558 SPD12118.1 OIVN01000958 SPC87933.1 OIVN01001202 SPC91100.1 OIVN01001091 SPC89740.1 OIVN01004420 SPD17230.1 KQ484186 KYP36645.1 KQ483702 KYP42748.1 CM003607 KYP67664.1 OIVN01001022 SPC88819.1 KQ483417 KYP53032.1 OIVN01000043 SPC73144.1 OIVN01000230 SPC76722.1 LBMM01007399 KMQ89763.1 OIVN01001309 SPC92453.1 OIVN01003498 SPD11675.1 OIVN01004603 SPD18338.1 OIVN01004456 SPD17553.1 OIVN01005059 SPD20680.1 OIVN01005186 SPD21283.1 OIVN01000657 SPC83575.1 OIVN01003902 SPD14305.1 AM438367 CAN71759.1 DF973332 GAU26309.1 ASHM01026402 PNX73691.1 AM442619 CAN74029.1 OIVN01006300 SPD30079.1 LVZM01023578 OUC39908.1 LBMM01011114 KMQ87021.1 LBMM01008978 KMQ88491.1 KQ483952 KYP38949.1 KF303287 AGW47867.1 GEZM01098152 JAV53972.1 AWWV01014897 OMO54668.1 AM451478 CAN80930.1 AM464898 CAN74767.1 ABLF02042028 DF973357 GAU27529.1 AM468226 CAN68235.1 OIVN01006381 SPD31869.1 LBMM01012321 KMQ86283.1 AAZX01006115 AAZX01021021 OIVN01001383 SPC93339.1 OIVN01002503 SPD04186.1 AWWV01013476 OMO61385.1 OIVN01003685 SPD12938.1 KQ483608 KYP44928.1 CM003609 KYP64079.1 DF974342 GAU47720.1 DF973742 GAU39052.1 OIVN01003578 SPD12232.1 CM007891 OTG34868.1 GEZM01048830 JAV76370.1 GEZM01017327 JAV90764.1
GEZM01082909 JAV61231.1 ABLF02062902 ABLF02031072 ABLF02059092 ABLF02060653 ABLF02023757 ABLF02028466 ABLF02059696 ABLF02055865 LBMM01012645 KMQ86082.1 ABLF02014335 ABLF02060220 GBHO01019780 JAG23824.1 GBHO01019781 JAG23823.1 GDHC01011760 JAQ06869.1 FJ544851 ACL97383.1 OIVN01002812 SPD06621.1 KQ484038 KYP38095.1 AGCT01017703 KYP77285.1 KL367580 KFD63098.1 FJ544856 ACL97386.1 OIVN01006160 SPD26619.1 KQ460882 KPJ11482.1 FJ544853 ACL97384.1 KQ483680 KYP43165.1 OIVN01000147 SPC75223.1 OIVN01001850 SPC98251.1 OIVN01002946 SPD07686.1 OIVN01000089 SPC74041.1 OIVN01003423 SPD11099.1 OIVN01004957 SPD20159.1 OIVN01006462 SPD33594.1 OIVN01002925 SPD07540.1 DF973531 GAU33542.1 OIVN01004307 SPD16591.1 OIVN01000823 SPC86071.1 OIVN01000418 SPC79865.1 OIVN01006125 SPD25623.1 JYDJ01000099 KRX44323.1 OIVN01005591 SPD23220.1 ASHM01021274 PNY02378.1 OIVN01005657 SPD23465.1 OIVN01003558 SPD12118.1 OIVN01000958 SPC87933.1 OIVN01001202 SPC91100.1 OIVN01001091 SPC89740.1 OIVN01004420 SPD17230.1 KQ484186 KYP36645.1 KQ483702 KYP42748.1 CM003607 KYP67664.1 OIVN01001022 SPC88819.1 KQ483417 KYP53032.1 OIVN01000043 SPC73144.1 OIVN01000230 SPC76722.1 LBMM01007399 KMQ89763.1 OIVN01001309 SPC92453.1 OIVN01003498 SPD11675.1 OIVN01004603 SPD18338.1 OIVN01004456 SPD17553.1 OIVN01005059 SPD20680.1 OIVN01005186 SPD21283.1 OIVN01000657 SPC83575.1 OIVN01003902 SPD14305.1 AM438367 CAN71759.1 DF973332 GAU26309.1 ASHM01026402 PNX73691.1 AM442619 CAN74029.1 OIVN01006300 SPD30079.1 LVZM01023578 OUC39908.1 LBMM01011114 KMQ87021.1 LBMM01008978 KMQ88491.1 KQ483952 KYP38949.1 KF303287 AGW47867.1 GEZM01098152 JAV53972.1 AWWV01014897 OMO54668.1 AM451478 CAN80930.1 AM464898 CAN74767.1 ABLF02042028 DF973357 GAU27529.1 AM468226 CAN68235.1 OIVN01006381 SPD31869.1 LBMM01012321 KMQ86283.1 AAZX01006115 AAZX01021021 OIVN01001383 SPC93339.1 OIVN01002503 SPD04186.1 AWWV01013476 OMO61385.1 OIVN01003685 SPD12938.1 KQ483608 KYP44928.1 CM003609 KYP64079.1 DF974342 GAU47720.1 DF973742 GAU39052.1 OIVN01003578 SPD12232.1 CM007891 OTG34868.1 GEZM01048830 JAV76370.1 GEZM01017327 JAV90764.1
Proteomes
Pfam
Interpro
IPR001584
Integrase_cat-core
+ More
IPR039537 Retrotran_Ty1/copia-like
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR013103 RVT_2
IPR025724 GAG-pre-integrase_dom
IPR036397 RNaseH_sf
IPR025314 DUF4219
IPR036965 Terpene_synth_N_sf
IPR008930 Terpenoid_cyclase/PrenylTrfase
IPR008949 Isoprenoid_synthase_dom_sf
IPR005630 Terpene_synthase_metal-bd
IPR034741 Terpene_cyclase-like_1_C
IPR001906 Terpene_synth_N
IPR041577 RT_RNaseH_2
IPR005123 Oxoglu/Fe-dep_dioxygenase
IPR027443 IPNS-like
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR023346 Lysozyme-like_dom_sf
IPR000726 Glyco_hydro_19_cat
IPR033133 PUM-HD
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR001313 Pumilio_RNA-bd_rpt
IPR029057 PRTase-like
IPR001357 BRCT_dom
IPR036420 BRCT_dom_sf
IPR032675 LRR_dom_sf
IPR021720 Malectin_dom
IPR017441 Protein_kinase_ATP_BS
IPR013083 Znf_RING/FYVE/PHD
IPR039537 Retrotran_Ty1/copia-like
IPR036875 Znf_CCHC_sf
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR013103 RVT_2
IPR025724 GAG-pre-integrase_dom
IPR036397 RNaseH_sf
IPR025314 DUF4219
IPR036965 Terpene_synth_N_sf
IPR008930 Terpenoid_cyclase/PrenylTrfase
IPR008949 Isoprenoid_synthase_dom_sf
IPR005630 Terpene_synthase_metal-bd
IPR034741 Terpene_cyclase-like_1_C
IPR001906 Terpene_synth_N
IPR041577 RT_RNaseH_2
IPR005123 Oxoglu/Fe-dep_dioxygenase
IPR027443 IPNS-like
IPR011009 Kinase-like_dom_sf
IPR001245 Ser-Thr/Tyr_kinase_cat_dom
IPR008271 Ser/Thr_kinase_AS
IPR000719 Prot_kinase_dom
IPR023346 Lysozyme-like_dom_sf
IPR000726 Glyco_hydro_19_cat
IPR033133 PUM-HD
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR001313 Pumilio_RNA-bd_rpt
IPR029057 PRTase-like
IPR001357 BRCT_dom
IPR036420 BRCT_dom_sf
IPR032675 LRR_dom_sf
IPR021720 Malectin_dom
IPR017441 Protein_kinase_ATP_BS
IPR013083 Znf_RING/FYVE/PHD
SUPFAM
Gene 3D
ProteinModelPortal
V5GAG2
A0A1Y1NAZ7
A0A1Y1N668
A0A1Y1KL90
X1X3E5
X1X2J4
+ More
J9L0N0 X1WZA6 A0A0J7K6W1 X1WT18 A0A0A9XV68 A0A0A9XY84 A0A146LH62 B8YLY3 A0A2N9H4R5 A0A151R6U8 A0A151UDA6 A0A085N0V2 B8YLY6 A0A2N9IR42 A0A0N1ICY1 B8YLY4 A0A151RL25 A0A2N9EK88 A0A2N9GG47 A0A2N9GZK0 A0A2N9EH79 A0A2N9HHJ9 A0A2N9I807 A0A2N9JAE7 A0A2N9H7C1 A0A2Z6N9Q0 A0A2N9HY48 A0A2N9FGL6 A0A2N9EYD8 A0A2N9ING1 A0A0V0U113 A0A2N9IGI9 A0A2K3NH55 A0A2N9IGN4 A0A2N9HJF5 A0A2N9FL83 A0A2N9FVE8 A0A2N9FRN5 A0A2N9HZR4 A0A151R2G6 A0A151RJN4 A0A151TKT1 A0A2N9FPD9 A0A151SDY6 A0A2N9EER7 A0A2N9EPH6 A0A0J7KH47 A0A2N9FZ01 A0A2N9HIB6 A0A2N9I1P0 A0A2N9I0K6 A0A2N9I9F7 A0A2N9IB18 A0A2N9F910 A0A2N9HQS0 A5AWP3 A0A2Z6N4D2 A0A2K3L574 A5B0R8 A0A2N9J0X3 A0A1Y3E9R1 A0A0J7K9V7 A0A0J7KE66 A0A151R8W9 A0A059QBK0 A0A1Y1JXD5 A0A1R3G990 A5B9M8 A5BMR0 X1XSG1 A0A2Z6N7E5 A5BR97 A0A2N9J5Y1 A0A0J7K724 K7JAV0 A0A2N9G211 A0A2N9GNZ7 A0A1R3GTE9 A0A3P1N2J7 A0A2N9HLW0 A0A151RQW4 A0A1U8MHG7 A0A151TAK4 A0A2Z6PCB3 A0A2Z6NRJ0 A0A2N9HJ44 A0A251VGU8 A0A1Y1LVN7 A0A1Y1MYP5
J9L0N0 X1WZA6 A0A0J7K6W1 X1WT18 A0A0A9XV68 A0A0A9XY84 A0A146LH62 B8YLY3 A0A2N9H4R5 A0A151R6U8 A0A151UDA6 A0A085N0V2 B8YLY6 A0A2N9IR42 A0A0N1ICY1 B8YLY4 A0A151RL25 A0A2N9EK88 A0A2N9GG47 A0A2N9GZK0 A0A2N9EH79 A0A2N9HHJ9 A0A2N9I807 A0A2N9JAE7 A0A2N9H7C1 A0A2Z6N9Q0 A0A2N9HY48 A0A2N9FGL6 A0A2N9EYD8 A0A2N9ING1 A0A0V0U113 A0A2N9IGI9 A0A2K3NH55 A0A2N9IGN4 A0A2N9HJF5 A0A2N9FL83 A0A2N9FVE8 A0A2N9FRN5 A0A2N9HZR4 A0A151R2G6 A0A151RJN4 A0A151TKT1 A0A2N9FPD9 A0A151SDY6 A0A2N9EER7 A0A2N9EPH6 A0A0J7KH47 A0A2N9FZ01 A0A2N9HIB6 A0A2N9I1P0 A0A2N9I0K6 A0A2N9I9F7 A0A2N9IB18 A0A2N9F910 A0A2N9HQS0 A5AWP3 A0A2Z6N4D2 A0A2K3L574 A5B0R8 A0A2N9J0X3 A0A1Y3E9R1 A0A0J7K9V7 A0A0J7KE66 A0A151R8W9 A0A059QBK0 A0A1Y1JXD5 A0A1R3G990 A5B9M8 A5BMR0 X1XSG1 A0A2Z6N7E5 A5BR97 A0A2N9J5Y1 A0A0J7K724 K7JAV0 A0A2N9G211 A0A2N9GNZ7 A0A1R3GTE9 A0A3P1N2J7 A0A2N9HLW0 A0A151RQW4 A0A1U8MHG7 A0A151TAK4 A0A2Z6PCB3 A0A2Z6NRJ0 A0A2N9HJ44 A0A251VGU8 A0A1Y1LVN7 A0A1Y1MYP5
Ontologies
KEGG
GO
PANTHER
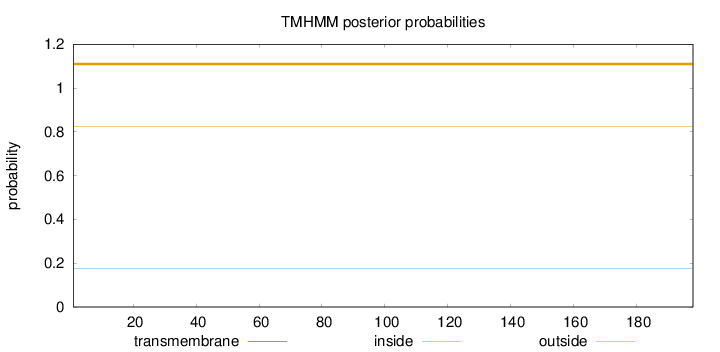
Topology
Length:
198
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.17700
outside
1 - 198
Population Genetic Test Statistics
Pi
0.6722
Theta
1.906434
Tajima's D
-1.386237
CLR
0.318539
CSRT
0.0653467326633668
Interpretation
Uncertain
