Gene
KWMTBOMO01983
Pre Gene Modal
BGIBMGA005909
Annotation
PREDICTED:_ankyrin-2_isoform_X1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.507 Nuclear Reliability : 1.251
Sequence
CDS
ATGGAAAATTCTTTAGAAGAAGCTTTATTTCAAGCTATCCTTCTTGAAGATCAGGGCTCAATCTCGCACTTATTATCAAAAGGGGCTAATCCAAATAAGACAGCAAGTCGTGGAAAGACATGCTTAGGCGAAGCTGCTAACACAGGAAATGTTGACGTAATGAAACTTCTCATAACAGCCAGCAAAACACCACAAAGTAAGAATAACCAAGCACCTTCAAGAAAAAGGACGTTAAAATGCCATAAAAGAAAGTTCAAACATGATGGACAAATAAGTGAAACAGTTGTGAAATTTAAAAACATTAATGAGAAAAAAATCAAAGATTATAGTGTAGGAAAATGTGAAAAATTAGATTTATCTCACACTGGACAAATTGAAAAAAATAAAGGATATTTTGTGTTTATACACAACGACGGAAGTAGTAGTGATGAAACACGCGTTCCTAGTTTGAAATCTCCGAAAAGCTCATCATCTATGACTCCTTCACCTCAAGCAGACTTGGAGTGGGATGAAGATATAGGCAATACTGCTCCAACTACCAGTGAAGATGAAACATGGTCATCAATGTATAAGTGGTATGCAGCAATATTAGAATCCACAGGGGCTGCAATAGCATCAGCTTCAAAAGTAACTAATGGCATAGACCAGCAAGATGCCTTTATGAGAACTGCCCTACATTATGCTTCAGAACAGGGACATATTGATACTGCAAAGCTTTTAATAGAAGCTGGATGTAAAATAGATGTTACAGCAAGTGATGGCCTGACACCTTTACATGTAGCAGTTGTTAAGAATCATATAGAAATTGTGAGACTACTTATTGCTGCTGGAAGTAATGTAAACTACAAAACTCACGAAAAAATGACTCCATTACACTTTGCATCTTCAAGAGGATTTTTAGATATTGTTAAATTATTAGTCAGTAATGGAGCACTTTTAGAAGCAAGAGACACAGCTGAAAGAACTGCATTATATCTAGCAGCAGGAAGGGGCCACTGTGATGTTGTCAAATACCTAATTCATAGCGGAGCCAATGTTAATGGTGAAGAAATTCATGGTTATACACCCCTATGTGAAGCAGTATGGCAAAGATATACGACGGTTGTCGAAATTTTACTATTAGCAGGCGCTCGCATCACACACTCGCATAGACTTTTGCATAATGCAATTATACAAAGACAGGAACCTATAGTTAAGATGCTAACTAAAATCGGTGGAGGTATAAATCTACACAATGACAATGGGGACACACCCCTACTTTTGGCGGCAAGATTATCGCAGCCTTCAGTTGCTAGATTATTGCTTGAGAAAGGTGCTAATGTAAACTCTTGCAACAATATCACGGGTGCTAATGCATTACATATAGCAGTTGAAAGCGTGGAATCACCTGAACATTTTGAAGTTCTATTGAAATGTTTATTGGATTATAATATAGATATGAACGCTATTACATTAATAGGCGACACAGCTTTAAACAGAGCTCTTTTGCTACAAAAGGATCATGCCGCAATTTTGTTGATTCGGTATGGAGCCGATGTTAATACGTGTGATTTACATTCATGCGGTTTGGACAATTTGTCAATAGCTAGCAGGAGAGAGACGCACGTTTTAGCGCGGATGTTGCTCAAGGCGGGGCATTACGTGATACCTCAAGATCCTAGTATTACTACTAAATCAACATGCACTACAAATTGGGTACAACAGACTTGTAAACAGCCCTTGACATTATCAGATTATTGTAGGATAACAATAAGGCGTATGTGTAAAAGTAATAAATTATTTCGTTTTGTGGCCACGTTACCTTTACCCCGTGGTTTAAAAAGGTTCCTCATGATGGAAGACGAGGGGCTCGAATAA
Protein
MENSLEEALFQAILLEDQGSISHLLSKGANPNKTASRGKTCLGEAANTGNVDVMKLLITASKTPQSKNNQAPSRKRTLKCHKRKFKHDGQISETVVKFKNINEKKIKDYSVGKCEKLDLSHTGQIEKNKGYFVFIHNDGSSSDETRVPSLKSPKSSSSMTPSPQADLEWDEDIGNTAPTTSEDETWSSMYKWYAAILESTGAAIASASKVTNGIDQQDAFMRTALHYASEQGHIDTAKLLIEAGCKIDVTASDGLTPLHVAVVKNHIEIVRLLIAAGSNVNYKTHEKMTPLHFASSRGFLDIVKLLVSNGALLEARDTAERTALYLAAGRGHCDVVKYLIHSGANVNGEEIHGYTPLCEAVWQRYTTVVEILLLAGARITHSHRLLHNAIIQRQEPIVKMLTKIGGGINLHNDNGDTPLLLAARLSQPSVARLLLEKGANVNSCNNITGANALHIAVESVESPEHFEVLLKCLLDYNIDMNAITLIGDTALNRALLLQKDHAAILLIRYGADVNTCDLHSCGLDNLSIASRRETHVLARMLLKAGHYVIPQDPSITTKSTCTTNWVQQTCKQPLTLSDYCRITIRRMCKSNKLFRFVATLPLPRGLKRFLMMEDEGLE
Summary
Uniprot
H9J8R7
A0A2A4K1M0
A0A2W1BFY5
A0A194PLD6
A0A2H1WNH6
A0A212ELS1
+ More
A0A067RNN4 A0A2J7RF50 A0A1B6E1A9 E0VRA7 A0A0L7L3V3 A0A224XH07 A0A023EY14 A0A146LDH9 A0A0K8SLG9 A0A1Y1M308 D6WMU1 A0A1W4WZX8 A0A0A9YCF6 T1H8H4 A0A0T6BAC2 A0A146M7C6 A0A2P8ZM64 N6U9G1 A0A1L8DIU6 A0A1L8DJ22 W4ZFM7 W4XCZ4 W8B511 W4XDD0 A2FZX0
A0A067RNN4 A0A2J7RF50 A0A1B6E1A9 E0VRA7 A0A0L7L3V3 A0A224XH07 A0A023EY14 A0A146LDH9 A0A0K8SLG9 A0A1Y1M308 D6WMU1 A0A1W4WZX8 A0A0A9YCF6 T1H8H4 A0A0T6BAC2 A0A146M7C6 A0A2P8ZM64 N6U9G1 A0A1L8DIU6 A0A1L8DJ22 W4ZFM7 W4XCZ4 W8B511 W4XDD0 A2FZX0
Pubmed
EMBL
BABH01040796
NWSH01000276
PCG77828.1
KZ150090
PZC73698.1
KQ459601
+ More
KPI93808.1 ODYU01009901 SOQ54623.1 AGBW02013994 OWR42419.1 KK852514 KDR22205.1 NEVH01004414 PNF39440.1 GEDC01025756 GEDC01005598 JAS11542.1 JAS31700.1 DS235459 EEB15913.1 JTDY01003128 KOB70100.1 GFTR01007328 JAW09098.1 GBBI01004739 JAC13973.1 GDHC01014059 JAQ04570.1 GBRD01016228 GBRD01011663 JAG54161.1 GEZM01042428 JAV79861.1 KQ971342 EFA03060.1 GBHO01042875 GBHO01014822 GBHO01014821 GBHO01014820 GBHO01014819 GBHO01014817 GBHO01014816 GBHO01008019 GBHO01008018 GBHO01008017 GBHO01008016 GBHO01008014 GBHO01008013 GBHO01004004 GBHO01002172 GBHO01002157 GBRD01011664 GBRD01011662 GDHC01013018 GDHC01004428 JAG00729.1 JAG28782.1 JAG28783.1 JAG28784.1 JAG28785.1 JAG28787.1 JAG28788.1 JAG35585.1 JAG35586.1 JAG35587.1 JAG35588.1 JAG35590.1 JAG35591.1 JAG39600.1 JAG41432.1 JAG41447.1 JAG54160.1 JAQ05611.1 JAQ14201.1 ACPB03005859 LJIG01002672 KRT84268.1 GDHC01002966 JAQ15663.1 PYGN01000018 PSN57585.1 APGK01037237 APGK01037238 APGK01037239 KB740948 ENN77286.1 GFDF01007722 JAV06362.1 GFDF01007709 JAV06375.1 AAGJ04102176 AAGJ04165197 GAMC01010320 JAB96235.1 AAGJ04091367 DS114190 EAX89549.1
KPI93808.1 ODYU01009901 SOQ54623.1 AGBW02013994 OWR42419.1 KK852514 KDR22205.1 NEVH01004414 PNF39440.1 GEDC01025756 GEDC01005598 JAS11542.1 JAS31700.1 DS235459 EEB15913.1 JTDY01003128 KOB70100.1 GFTR01007328 JAW09098.1 GBBI01004739 JAC13973.1 GDHC01014059 JAQ04570.1 GBRD01016228 GBRD01011663 JAG54161.1 GEZM01042428 JAV79861.1 KQ971342 EFA03060.1 GBHO01042875 GBHO01014822 GBHO01014821 GBHO01014820 GBHO01014819 GBHO01014817 GBHO01014816 GBHO01008019 GBHO01008018 GBHO01008017 GBHO01008016 GBHO01008014 GBHO01008013 GBHO01004004 GBHO01002172 GBHO01002157 GBRD01011664 GBRD01011662 GDHC01013018 GDHC01004428 JAG00729.1 JAG28782.1 JAG28783.1 JAG28784.1 JAG28785.1 JAG28787.1 JAG28788.1 JAG35585.1 JAG35586.1 JAG35587.1 JAG35588.1 JAG35590.1 JAG35591.1 JAG39600.1 JAG41432.1 JAG41447.1 JAG54160.1 JAQ05611.1 JAQ14201.1 ACPB03005859 LJIG01002672 KRT84268.1 GDHC01002966 JAQ15663.1 PYGN01000018 PSN57585.1 APGK01037237 APGK01037238 APGK01037239 KB740948 ENN77286.1 GFDF01007722 JAV06362.1 GFDF01007709 JAV06375.1 AAGJ04102176 AAGJ04165197 GAMC01010320 JAB96235.1 AAGJ04091367 DS114190 EAX89549.1
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J8R7
A0A2A4K1M0
A0A2W1BFY5
A0A194PLD6
A0A2H1WNH6
A0A212ELS1
+ More
A0A067RNN4 A0A2J7RF50 A0A1B6E1A9 E0VRA7 A0A0L7L3V3 A0A224XH07 A0A023EY14 A0A146LDH9 A0A0K8SLG9 A0A1Y1M308 D6WMU1 A0A1W4WZX8 A0A0A9YCF6 T1H8H4 A0A0T6BAC2 A0A146M7C6 A0A2P8ZM64 N6U9G1 A0A1L8DIU6 A0A1L8DJ22 W4ZFM7 W4XCZ4 W8B511 W4XDD0 A2FZX0
A0A067RNN4 A0A2J7RF50 A0A1B6E1A9 E0VRA7 A0A0L7L3V3 A0A224XH07 A0A023EY14 A0A146LDH9 A0A0K8SLG9 A0A1Y1M308 D6WMU1 A0A1W4WZX8 A0A0A9YCF6 T1H8H4 A0A0T6BAC2 A0A146M7C6 A0A2P8ZM64 N6U9G1 A0A1L8DIU6 A0A1L8DJ22 W4ZFM7 W4XCZ4 W8B511 W4XDD0 A2FZX0
PDB
6MOL
E-value=1.23921e-40,
Score=420
Ontologies
GO
Topology
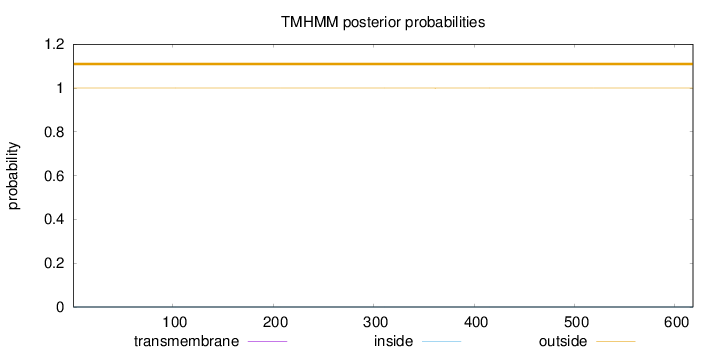
Length:
618
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00329
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00011
outside
1 - 618
Population Genetic Test Statistics
Pi
228.483377
Theta
173.780235
Tajima's D
1.150307
CLR
0.006159
CSRT
0.708464576771161
Interpretation
Uncertain
