Gene
KWMTBOMO01980
Pre Gene Modal
BGIBMGA005912
Annotation
hypothetical_protein_KGM_03031_[Danaus_plexippus]
Full name
Dipeptidase
Location in the cell
Mitochondrial Reliability : 1.592 Nuclear Reliability : 1.947
Sequence
CDS
ATGGTTGATGGAGAAGCCAGCGGCCACGGTGCTACGGTCGGAAACCCACCGTCGGATTGTGTTGATTGTGAAGCTTGCATAGCATTAGCTGCCCAGCGACAGTGTCTAAGTGAACACATATCAAAACATGATTTAGACTTAGTTTGCAATTGTAATATACAACAGAACAACTACCTAAACTGTCATCGAAATGTTTCTGATCATCGAGGCCACACTCTAAATATTTATGATTTAGAGAGAATCAAAAGAGAAACCGATAGAAGTCTAATAAACGTGGGATCACACCAAAGTCTCGACAGAAAACAAACAAGAAATAAATCCATTGGTGTCTCTAATTATATTTCGAAACAATTAATATCGTATGACCCAAGAAATACTCTTGTCCAAAAAATCAAGAGAAAACTATCTAATTTGAAAAAAATCGGCGGAGATGCTATCACCAAGAAACATCAAAACGCTGTCCAACTCGCCGATATAGCATCCGGGTCTACCAGCTCCCTCAAAAAACTATCTATGCTGTCGATTGCGGACAAACCACAAAGAAGAGTTAACAGCCATAGCAACATATATCCATCGACGACAGTAAATAGAGACGACTACAAACACAAAGCGTCAACATACGCAAATTTTTCTGGTAACGTGGACCAATGCTTTGATAGACTGAAAGATTCCAATACAGTCATGGATATAAACAATACGACACAAAAAATAAGAACTTTTGATAACAATCAGGAGCTGATCCGCAGGAATAATGTGGATATAGATGCAATGAAATTTTCTACTTTAGACAATCGAAGCAGATGTAGAAGTATCCGTAGACAACTATCATACAACGATTTTCCTAGTGATCGATGCAAGGAGAAAATCACGGAGACATGGATGACAAATGAAAATACTATTGAGTCCAGGAGTACTCGAAGTGATTTAGTCGACGATGATGTCCACACGTTTATTGACAACGACGTAGTGGCAATGACAGGAACGAGGCCGACACACGCACATTGTACCTGCACCCAGAGCTATGATAATAGAATGCCTTCTTTATTCTACGACTCGCAAGGCAAGAAACGGTCGGCGCCAGCACCTGACGTGACAGAGCGTAGTCAAGGGACTCGCGCAAACGTTTGTAGCGCTTGCAAAGTACCGCAATGGATATCTACTGGTACAACATCGACTACAAGCCACAGTTCAACTACAACTGGAAATTCTTCTCGTTTTTCAATATGGCACCGTCGATGGTGCTGCGTAGCAGTTTTAGTACTGGTAGCCGGGACCGCGTGCGTAGCAGGCCCTCTTGCTTTGAGAGCTCCTCCAGGCGCTCCGATGCATGAAAGACTTCGACTGGCCGAAAGACTATTACACGATACGCCTCTTATCGACGGACACAACGACTTGCCTTGGAATATAAGAAAATTTCTACACAACAAGATTAAAGACTTTTCGTTTGATGAGGACTTGAGGACGATATCACCATGGGCGACGAGCTCGTGGAGCCACACTGATTTGCCGAGGTTAAAACAAGGCAGGGTCGCTGCTCAGTTCTGGGCGGCGTACGTGCCTTGCGATGCGCAACACCGCGACGCCGTGCAGCTGACGTTCGAGCAAATTGACCTCATCCAGCGACTCACTGAAAAATATCAACCTCAGCTTACAATGTGCACTTCTGCCGAAGATATTAAAATAGCGCACGCTAATCATCGTGTATGCTCGTTGATTGGTGTCGAAGGCGGTCACGCTATTGGCAGTTCTCTTGGGGTGCTAAGAACACTGTACCAGGTCGGCGTTCGTTACTTGACATTGACCTCTACTTGTGACACGCCCTGGGCTGAGTGCGCTTCTGCTGACAGAGTTGAACCATCGCCCCGGGGAGGACTCACGGCTTTTGGAAAGGTAGTAATAAAGGAAATGAACAGATTGGGAATGCTAGTGGATTTATCTCACGTGTCAGAGCGAACTATGCGTGATGCTCTGGCTGTCTCACGAGCACCAGTGCTTTTTTCTCACTCTTCTGCACGAGCTCTTTGCAACGTTACTCGTAACGTTCCTGACACAATACTGAGAATGTTAGCTGCCAACAAAGGACTCATAATGGTGAATTTTTATACTTCATTCCTGACTTGTAAGGAAACAGCAACCGTTCAAGATGCGATTGCTCACATAAATCATATACGGAACGTGGCCGGTGTGGACAGCGTCGGCCTTGGCGCCGGGTACGATGGGATTAATTATACACCACAAGGTCTGGAAGATGTATCATCCTATCCATTGTTATTTGCCGAGCTGATGGAGGCAGGCTGGAGCATAGAAGATCTTCGTAAGCTTGCCGGACTGAATCTACTCCGGGTAATGGGCGCAACAGAGAGGGTAGCTAAAGAATTGGCAGCTGCTCGTGTTGTACCTTACGAAGACGTGCCACCGCGAATTTTCGACGCTCATAACTGCTCCAGTCAAGATGTTTAA
Protein
MVDGEASGHGATVGNPPSDCVDCEACIALAAQRQCLSEHISKHDLDLVCNCNIQQNNYLNCHRNVSDHRGHTLNIYDLERIKRETDRSLINVGSHQSLDRKQTRNKSIGVSNYISKQLISYDPRNTLVQKIKRKLSNLKKIGGDAITKKHQNAVQLADIASGSTSSLKKLSMLSIADKPQRRVNSHSNIYPSTTVNRDDYKHKASTYANFSGNVDQCFDRLKDSNTVMDINNTTQKIRTFDNNQELIRRNNVDIDAMKFSTLDNRSRCRSIRRQLSYNDFPSDRCKEKITETWMTNENTIESRSTRSDLVDDDVHTFIDNDVVAMTGTRPTHAHCTCTQSYDNRMPSLFYDSQGKKRSAPAPDVTERSQGTRANVCSACKVPQWISTGTTSTTSHSSTTTGNSSRFSIWHRRWCCVAVLVLVAGTACVAGPLALRAPPGAPMHERLRLAERLLHDTPLIDGHNDLPWNIRKFLHNKIKDFSFDEDLRTISPWATSSWSHTDLPRLKQGRVAAQFWAAYVPCDAQHRDAVQLTFEQIDLIQRLTEKYQPQLTMCTSAEDIKIAHANHRVCSLIGVEGGHAIGSSLGVLRTLYQVGVRYLTLTSTCDTPWAECASADRVEPSPRGGLTAFGKVVIKEMNRLGMLVDLSHVSERTMRDALAVSRAPVLFSHSSARALCNVTRNVPDTILRMLAANKGLIMVNFYTSFLTCKETATVQDAIAHINHIRNVAGVDSVGLGAGYDGINYTPQGLEDVSSYPLLFAELMEAGWSIEDLRKLAGLNLLRVMGATERVAKELAAARVVPYEDVPPRIFDAHNCSSQDV
Summary
Catalytic Activity
Hydrolysis of dipeptides.
Cofactor
Zn(2+)
Subunit
Homodimer; disulfide-linked.
Similarity
Belongs to the metallo-dependent hydrolases superfamily. Peptidase M19 family.
Uniprot
EC Number
3.4.13.19
EMBL
Proteomes
Pfam
PF01244 Peptidase_M19
SUPFAM
SSF51556
SSF51556
CDD
ProteinModelPortal
PDB
1ITU
E-value=3.13976e-89,
Score=841
Ontologies
GO
PANTHER
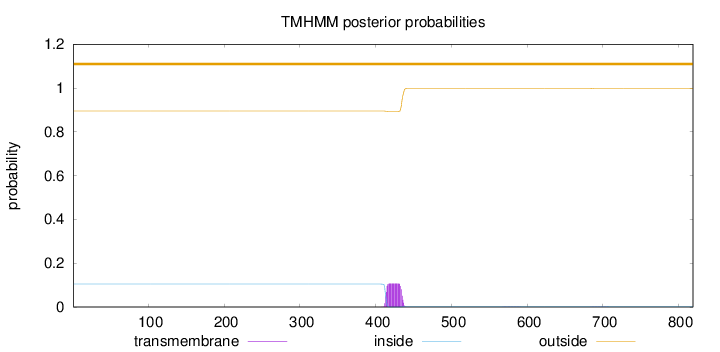
Topology
Subcellular location
Membrane
Length:
819
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.36746
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10477
outside
1 - 819
Population Genetic Test Statistics
Pi
278.525844
Theta
184.635232
Tajima's D
1.608109
CLR
0.005763
CSRT
0.807909604519774
Interpretation
Uncertain
