Gene
KWMTBOMO01979
Pre Gene Modal
BGIBMGA014036
Annotation
PREDICTED:_transcription_initiation_factor_TFIID_subunit_5_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.433 Extracellular Reliability : 1.176
Sequence
CDS
ATGAGTGAGAAATCAACCCCACTGCTGGCTGTTTTACAGTTACTTAGAAAGTACAATCTAAAGAGTACAGAAGACATTTTAAGAAAAGAAGCTAGTCTAGGAGACGTCGAATGTGAAAGCTTAGATCTTCCTGAAGTAGAACTAGCGAGCATTCTTACCGCTCATCACACTGAAAGCGACCCCTTTACGTACGAATTGGCGTACGACGGTCTAAAAAAATTTATTGAAAATTCTTTAGATTTGTACAAGTATGAATTGTCCACACTATTGTATCCTGTGTTTGTTCACATGTATCTTCTGTTGATACTGTATGATCATGTTGAACACGCTGTCAGTTTCTTGGAAAAGTTTGGAATAGAACAAGAAGATTACTATCAAGAGGACTTGAAAAAGTTGTCTATTGTAAAACATAAGGATCAGATAAAAGGAAATGAACTAGCAGAAATATACAGCACAAATAAATTTACAGTTCAGTTATCAAGAGATGCATCTTCACAACTGAAACGTTACTTACACGAACAGAAAAATTCCTCAGTAATAATAAATATAATAAATAATCATATACAAATCGAAGTCCATGATGGTCCAGGAAGAACTCAAGCACAAGTTCGAGCTACAGCTGGAGGGGTACTTGGAGAACCAATGAAAAGTGAAAATCGCACCAAAGTGTATTATGGGCTCCTGAAAGAGCCCGATATTCAAGTTCTACCGGCTCCGGTAGAAGACGAAGAAGAAGTAGAAGAAACCCCTGACAAACCAAAAAAGAAAAAAGCGAAAAAAGATAATCTTTTTATGAAGAAACCAAAATCTGACCCAAATGCACCTCCTAATGACAGGATCCCTCTACCAGATCTAAAAGAAATTGATAAAGTAGAGAAAGGCAAAGCACTCAGAGAAGCATCAAAACGTGTTCAGCTAGGACCAGACAGCTTGCCCTCTATTTGTTTCTATACATTACTGAACAGCGGACATACTGCAATTAGTTGTGATATTTGTGACGATTCCACCCTTTTGGCTGTGGGATTTAATAACTCAAACATAAAGGTATGGACATTGACTCCCATTAAACTGCGAGGAATGAAATCTGCAGAAAAACTACAAGATATAGACAGGGAAGCAGGTGATGTACTCGTGAGGATGATGGAGGAAAAGGACCGTGATACGTGCCGCAACCTTTTTGGGCACTCCGGCCCCGTTTATAAAGTTGCATTTGATCCTTTTAAAACAATGCTGCTTTCGTGCTCTGAGGATACAACTGTGAGATTGTGGTCACTACAGTGTTGGACGTGTCTTGTTGTGTACCGTGGACACGTATGGCCGGTCTGGGATGTGCGCTGGTCCCCTCATGGCCACTACTTTGCTTCCAGCGGCCATGATCGTACTGCTCGACTATGGGCGACTGATCACCACCTTCCACTGAGGATATTTATAGGACATTATTCGGATGTAGATTGTATTCAGTTCCACCCGAACTCAAATTATATAGCAACTGGATCTAGCGACAGAACAGTTCGACTGTGGGATTGTTTAACTGGTAGTCAAGTTCGAATAATGACAGGACATAAGTCAACCATTTTCACGTTGGCCTTTTCCGTTTGTGGAAGATGGATTGCTTCTGGTGGCAGTGGTGGAGATATGATGGTTTGGGATTTATCTACTGGGGGTCCAGCCGCTTCGTTACCCCCGGCACATACTGCTCCCATACACGCCTTAGCCTTCAGTCGTGATGGAACAGTTCTATCGTCAGGCTCATTGGATTGTACAATAAAATTGTGGGACTTCACAGCCATAACAGACGAATCTCCCATTGAAGAAACTCTAAATAACCCTATACCAAAAGAAGACAAATATCTATTACAATCATTTGCCACTAAAAGTTCCCCTGTAAAAGGCTTGCATTTTACAAGACGCAATTTACTACTTGCTGTTGGCACATATGAGAGCAGTGCTTAA
Protein
MSEKSTPLLAVLQLLRKYNLKSTEDILRKEASLGDVECESLDLPEVELASILTAHHTESDPFTYELAYDGLKKFIENSLDLYKYELSTLLYPVFVHMYLLLILYDHVEHAVSFLEKFGIEQEDYYQEDLKKLSIVKHKDQIKGNELAEIYSTNKFTVQLSRDASSQLKRYLHEQKNSSVIINIINNHIQIEVHDGPGRTQAQVRATAGGVLGEPMKSENRTKVYYGLLKEPDIQVLPAPVEDEEEVEETPDKPKKKKAKKDNLFMKKPKSDPNAPPNDRIPLPDLKEIDKVEKGKALREASKRVQLGPDSLPSICFYTLLNSGHTAISCDICDDSTLLAVGFNNSNIKVWTLTPIKLRGMKSAEKLQDIDREAGDVLVRMMEEKDRDTCRNLFGHSGPVYKVAFDPFKTMLLSCSEDTTVRLWSLQCWTCLVVYRGHVWPVWDVRWSPHGHYFASSGHDRTARLWATDHHLPLRIFIGHYSDVDCIQFHPNSNYIATGSSDRTVRLWDCLTGSQVRIMTGHKSTIFTLAFSVCGRWIASGGSGGDMMVWDLSTGGPAASLPPAHTAPIHALAFSRDGTVLSSGSLDCTIKLWDFTAITDESPIEETLNNPIPKEDKYLLQSFATKSSPVKGLHFTRRNLLLAVGTYESSA
Summary
Uniprot
H9JWX2
A0A2A4K1A0
A0A2H1X0B5
A0A2W1BHE8
A0A212ELQ3
A0A194RGS6
+ More
I4DKH6 A0A194PM50 A0A067QZS3 A0A2J7PLN0 A0A2J7PLN8 A0A1B0CJD6 A0A2A3ELK7 A0A088A129 A0A1L8DGY0 A0A1B6JIZ9 A0A0J7KDZ8 A0A232EF34 A0A1B6DEK0 E2C4F8 K7IZW9 A0A146MEU0 E2A7B5 A0A0V0G8P2 A0A182HFB3 A0A182N2H0 A0A084VIE5 F4WQV2 A0A182YJD1 Q173R0 A0A1Q3F438 A0A182PC89 A0A182T1V8 A0A1S4FFG7 A0A182R706 A0A182JTR5 A0A182M4W7 A0A158NTI8 E0VEZ5 B0WAT2 A0A182QCP7 A0A182WEI4 A0A151X3D8 A0A195EG06 A0A0M9A0M7 A0A151INR9 A0A336LNL9 A0A0P5U5C6 Q7PZ11 A0A0P5CSM7 A0A0P5U041 A0A182XKM2 A0A182V284 A0A182L4C0 A0A0P5XL53 A0A182TJL4 A0A182HGX8 A0A0N8A234 A0A182IVK7 E9FZR3 A0A0P6INE2 A0A1B6H3R1 A0A2M4AN40 W5JUP4 A0A3L8DJ74 A0A0P4XTX4 A0A0P5DHX6 A0A182FUT0 A0A195BRP9 A0A195ESN7 D6WNI9 T1HS24 A0A0C9RW60 A0A1Y1MMD9 A0A164UJL5 A0A1J1J8S1 A0A1B0DNX3 A0A0N8AC08 A0A026W853 A0A0L7QUA4 A0A3G2KX35 N6U9K1 A0A2J7PLN3 A0A154P2V5 A0A2L2YKU4 A0A3Q0JLC4 U4UGQ1 A0A2R5LLY4 A0A0A9YNJ7 B7QDX5 A0A131Y1H0 A0A131YK82 L7MLW4 A0A224Z1P1 A0A1I8QB31 A0A0P4W488 A0A1A9W6F2 A0A1A9ZHY3 A0A1B0BZC7
I4DKH6 A0A194PM50 A0A067QZS3 A0A2J7PLN0 A0A2J7PLN8 A0A1B0CJD6 A0A2A3ELK7 A0A088A129 A0A1L8DGY0 A0A1B6JIZ9 A0A0J7KDZ8 A0A232EF34 A0A1B6DEK0 E2C4F8 K7IZW9 A0A146MEU0 E2A7B5 A0A0V0G8P2 A0A182HFB3 A0A182N2H0 A0A084VIE5 F4WQV2 A0A182YJD1 Q173R0 A0A1Q3F438 A0A182PC89 A0A182T1V8 A0A1S4FFG7 A0A182R706 A0A182JTR5 A0A182M4W7 A0A158NTI8 E0VEZ5 B0WAT2 A0A182QCP7 A0A182WEI4 A0A151X3D8 A0A195EG06 A0A0M9A0M7 A0A151INR9 A0A336LNL9 A0A0P5U5C6 Q7PZ11 A0A0P5CSM7 A0A0P5U041 A0A182XKM2 A0A182V284 A0A182L4C0 A0A0P5XL53 A0A182TJL4 A0A182HGX8 A0A0N8A234 A0A182IVK7 E9FZR3 A0A0P6INE2 A0A1B6H3R1 A0A2M4AN40 W5JUP4 A0A3L8DJ74 A0A0P4XTX4 A0A0P5DHX6 A0A182FUT0 A0A195BRP9 A0A195ESN7 D6WNI9 T1HS24 A0A0C9RW60 A0A1Y1MMD9 A0A164UJL5 A0A1J1J8S1 A0A1B0DNX3 A0A0N8AC08 A0A026W853 A0A0L7QUA4 A0A3G2KX35 N6U9K1 A0A2J7PLN3 A0A154P2V5 A0A2L2YKU4 A0A3Q0JLC4 U4UGQ1 A0A2R5LLY4 A0A0A9YNJ7 B7QDX5 A0A131Y1H0 A0A131YK82 L7MLW4 A0A224Z1P1 A0A1I8QB31 A0A0P4W488 A0A1A9W6F2 A0A1A9ZHY3 A0A1B0BZC7
Pubmed
19121390
28756777
22118469
26354079
22651552
24845553
+ More
28648823 20798317 20075255 26823975 26483478 24438588 21719571 25244985 17510324 21347285 20566863 12364791 14747013 17210077 20966253 21292972 20920257 23761445 30249741 18362917 19820115 28004739 24508170 30329069 23537049 26561354 25401762 26830274 25576852 28797301
28648823 20798317 20075255 26823975 26483478 24438588 21719571 25244985 17510324 21347285 20566863 12364791 14747013 17210077 20966253 21292972 20920257 23761445 30249741 18362917 19820115 28004739 24508170 30329069 23537049 26561354 25401762 26830274 25576852 28797301
EMBL
BABH01042586
BABH01042587
BABH01042588
NWSH01000276
PCG77839.1
ODYU01012447
+ More
SOQ58751.1 KZ150090 PZC73691.1 AGBW02013994 OWR42424.1 KQ460398 KPJ15131.1 AK401794 BAM18416.1 KQ459601 KPI93814.1 KK853149 KDR10646.1 NEVH01024424 PNF17244.1 PNF17242.1 AJWK01014499 KZ288215 PBC32645.1 GFDF01008365 JAV05719.1 GECU01033538 GECU01026235 GECU01008536 JAS74168.1 JAS81471.1 JAS99170.1 LBMM01009068 KMQ88416.1 NNAY01005138 OXU16957.1 GEDC01013228 JAS24070.1 GL452462 EFN77195.1 GDHC01001117 JAQ17512.1 GL437329 EFN70612.1 GECL01002487 JAP03637.1 JXUM01036744 KQ561105 KXJ79684.1 ATLV01013354 KE524854 KFB37739.1 GL888275 EGI63465.1 CH477418 EAT41277.1 GFDL01012724 JAV22321.1 AXCM01001816 ADTU01025973 DS235100 EEB11969.1 DS231875 EDS41798.1 AXCN02001071 AXCN02001072 KQ982562 KYQ54901.1 KQ978983 KYN26807.1 KQ435776 KOX74930.1 KQ976919 KYN07085.1 UFQT01000092 SSX19596.1 GDIP01147593 GDIP01121720 JAL81994.1 AAAB01008986 EAA00089.4 GDIP01167381 JAJ56021.1 GDIP01119628 JAL84086.1 GDIP01070449 JAM33266.1 APCN01002367 GDIP01190065 JAJ33337.1 GL732528 EFX87221.1 GDIQ01002871 JAN91866.1 GECZ01000466 JAS69303.1 GGFK01008870 MBW42191.1 ADMH02000317 ETN66998.1 QOIP01000007 RLU20223.1 GDIP01240827 JAI82574.1 GDIP01157236 JAJ66166.1 KQ976424 KYM88616.1 KQ981993 KYN30927.1 KQ971343 EFA04573.1 ACPB03003000 GBYB01005128 GBYB01012006 JAG74895.1 JAG81773.1 GEZM01030354 GEZM01030349 JAV85605.1 LRGB01001581 KZS11419.1 CVRI01000070 CRL07321.1 AJVK01007807 AJVK01007808 GDIP01162273 JAJ61129.1 KK107347 EZA52240.1 KQ414735 KOC62223.1 MH712878 AYN64385.1 APGK01037418 KB740948 ENN77326.1 PNF17243.1 KQ434792 KZC05460.1 IAAA01035012 LAA08729.1 KB632288 ERL91528.1 GGLE01006414 MBY10540.1 GBHO01010936 GBRD01010238 JAG32668.1 JAG55586.1 ABJB010754237 ABJB010896936 ABJB010930261 DS916876 EEC17047.1 GEFM01003920 JAP71876.1 GEDV01009707 JAP78850.1 GACK01000845 JAA64189.1 GFPF01012622 MAA23768.1 GDRN01073399 JAI63398.1 JXJN01023092
SOQ58751.1 KZ150090 PZC73691.1 AGBW02013994 OWR42424.1 KQ460398 KPJ15131.1 AK401794 BAM18416.1 KQ459601 KPI93814.1 KK853149 KDR10646.1 NEVH01024424 PNF17244.1 PNF17242.1 AJWK01014499 KZ288215 PBC32645.1 GFDF01008365 JAV05719.1 GECU01033538 GECU01026235 GECU01008536 JAS74168.1 JAS81471.1 JAS99170.1 LBMM01009068 KMQ88416.1 NNAY01005138 OXU16957.1 GEDC01013228 JAS24070.1 GL452462 EFN77195.1 GDHC01001117 JAQ17512.1 GL437329 EFN70612.1 GECL01002487 JAP03637.1 JXUM01036744 KQ561105 KXJ79684.1 ATLV01013354 KE524854 KFB37739.1 GL888275 EGI63465.1 CH477418 EAT41277.1 GFDL01012724 JAV22321.1 AXCM01001816 ADTU01025973 DS235100 EEB11969.1 DS231875 EDS41798.1 AXCN02001071 AXCN02001072 KQ982562 KYQ54901.1 KQ978983 KYN26807.1 KQ435776 KOX74930.1 KQ976919 KYN07085.1 UFQT01000092 SSX19596.1 GDIP01147593 GDIP01121720 JAL81994.1 AAAB01008986 EAA00089.4 GDIP01167381 JAJ56021.1 GDIP01119628 JAL84086.1 GDIP01070449 JAM33266.1 APCN01002367 GDIP01190065 JAJ33337.1 GL732528 EFX87221.1 GDIQ01002871 JAN91866.1 GECZ01000466 JAS69303.1 GGFK01008870 MBW42191.1 ADMH02000317 ETN66998.1 QOIP01000007 RLU20223.1 GDIP01240827 JAI82574.1 GDIP01157236 JAJ66166.1 KQ976424 KYM88616.1 KQ981993 KYN30927.1 KQ971343 EFA04573.1 ACPB03003000 GBYB01005128 GBYB01012006 JAG74895.1 JAG81773.1 GEZM01030354 GEZM01030349 JAV85605.1 LRGB01001581 KZS11419.1 CVRI01000070 CRL07321.1 AJVK01007807 AJVK01007808 GDIP01162273 JAJ61129.1 KK107347 EZA52240.1 KQ414735 KOC62223.1 MH712878 AYN64385.1 APGK01037418 KB740948 ENN77326.1 PNF17243.1 KQ434792 KZC05460.1 IAAA01035012 LAA08729.1 KB632288 ERL91528.1 GGLE01006414 MBY10540.1 GBHO01010936 GBRD01010238 JAG32668.1 JAG55586.1 ABJB010754237 ABJB010896936 ABJB010930261 DS916876 EEC17047.1 GEFM01003920 JAP71876.1 GEDV01009707 JAP78850.1 GACK01000845 JAA64189.1 GFPF01012622 MAA23768.1 GDRN01073399 JAI63398.1 JXJN01023092
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000027135
+ More
UP000235965 UP000092461 UP000242457 UP000005203 UP000036403 UP000215335 UP000008237 UP000002358 UP000000311 UP000069940 UP000249989 UP000075884 UP000030765 UP000007755 UP000076408 UP000008820 UP000075885 UP000075901 UP000075900 UP000075881 UP000075883 UP000005205 UP000009046 UP000002320 UP000075886 UP000075920 UP000075809 UP000078492 UP000053105 UP000078542 UP000007062 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000075880 UP000000305 UP000000673 UP000279307 UP000069272 UP000078540 UP000078541 UP000007266 UP000015103 UP000076858 UP000183832 UP000092462 UP000053097 UP000053825 UP000019118 UP000076502 UP000079169 UP000030742 UP000001555 UP000095300 UP000091820 UP000092445 UP000092460
UP000235965 UP000092461 UP000242457 UP000005203 UP000036403 UP000215335 UP000008237 UP000002358 UP000000311 UP000069940 UP000249989 UP000075884 UP000030765 UP000007755 UP000076408 UP000008820 UP000075885 UP000075901 UP000075900 UP000075881 UP000075883 UP000005205 UP000009046 UP000002320 UP000075886 UP000075920 UP000075809 UP000078492 UP000053105 UP000078542 UP000007062 UP000076407 UP000075903 UP000075882 UP000075902 UP000075840 UP000075880 UP000000305 UP000000673 UP000279307 UP000069272 UP000078540 UP000078541 UP000007266 UP000015103 UP000076858 UP000183832 UP000092462 UP000053097 UP000053825 UP000019118 UP000076502 UP000079169 UP000030742 UP000001555 UP000095300 UP000091820 UP000092445 UP000092460
PRIDE
Pfam
Interpro
IPR017986
WD40_repeat_dom
+ More
IPR037264 TFIID_NTD2_sf
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR037783 Taf5
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR007582 TFIID_NTD2
IPR020472 G-protein_beta_WD-40_rep
IPR006594 LisH
IPR038273 Ndc80_sf
IPR005550 Kinetochore_Ndc80
IPR018556 DUF2013
IPR036028 SH3-like_dom_sf
IPR035514 SPIN90_SH3
IPR001452 SH3_domain
IPR037264 TFIID_NTD2_sf
IPR001680 WD40_repeat
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR037783 Taf5
IPR019775 WD40_repeat_CS
IPR036322 WD40_repeat_dom_sf
IPR007582 TFIID_NTD2
IPR020472 G-protein_beta_WD-40_rep
IPR006594 LisH
IPR038273 Ndc80_sf
IPR005550 Kinetochore_Ndc80
IPR018556 DUF2013
IPR036028 SH3-like_dom_sf
IPR035514 SPIN90_SH3
IPR001452 SH3_domain
Gene 3D
ProteinModelPortal
H9JWX2
A0A2A4K1A0
A0A2H1X0B5
A0A2W1BHE8
A0A212ELQ3
A0A194RGS6
+ More
I4DKH6 A0A194PM50 A0A067QZS3 A0A2J7PLN0 A0A2J7PLN8 A0A1B0CJD6 A0A2A3ELK7 A0A088A129 A0A1L8DGY0 A0A1B6JIZ9 A0A0J7KDZ8 A0A232EF34 A0A1B6DEK0 E2C4F8 K7IZW9 A0A146MEU0 E2A7B5 A0A0V0G8P2 A0A182HFB3 A0A182N2H0 A0A084VIE5 F4WQV2 A0A182YJD1 Q173R0 A0A1Q3F438 A0A182PC89 A0A182T1V8 A0A1S4FFG7 A0A182R706 A0A182JTR5 A0A182M4W7 A0A158NTI8 E0VEZ5 B0WAT2 A0A182QCP7 A0A182WEI4 A0A151X3D8 A0A195EG06 A0A0M9A0M7 A0A151INR9 A0A336LNL9 A0A0P5U5C6 Q7PZ11 A0A0P5CSM7 A0A0P5U041 A0A182XKM2 A0A182V284 A0A182L4C0 A0A0P5XL53 A0A182TJL4 A0A182HGX8 A0A0N8A234 A0A182IVK7 E9FZR3 A0A0P6INE2 A0A1B6H3R1 A0A2M4AN40 W5JUP4 A0A3L8DJ74 A0A0P4XTX4 A0A0P5DHX6 A0A182FUT0 A0A195BRP9 A0A195ESN7 D6WNI9 T1HS24 A0A0C9RW60 A0A1Y1MMD9 A0A164UJL5 A0A1J1J8S1 A0A1B0DNX3 A0A0N8AC08 A0A026W853 A0A0L7QUA4 A0A3G2KX35 N6U9K1 A0A2J7PLN3 A0A154P2V5 A0A2L2YKU4 A0A3Q0JLC4 U4UGQ1 A0A2R5LLY4 A0A0A9YNJ7 B7QDX5 A0A131Y1H0 A0A131YK82 L7MLW4 A0A224Z1P1 A0A1I8QB31 A0A0P4W488 A0A1A9W6F2 A0A1A9ZHY3 A0A1B0BZC7
I4DKH6 A0A194PM50 A0A067QZS3 A0A2J7PLN0 A0A2J7PLN8 A0A1B0CJD6 A0A2A3ELK7 A0A088A129 A0A1L8DGY0 A0A1B6JIZ9 A0A0J7KDZ8 A0A232EF34 A0A1B6DEK0 E2C4F8 K7IZW9 A0A146MEU0 E2A7B5 A0A0V0G8P2 A0A182HFB3 A0A182N2H0 A0A084VIE5 F4WQV2 A0A182YJD1 Q173R0 A0A1Q3F438 A0A182PC89 A0A182T1V8 A0A1S4FFG7 A0A182R706 A0A182JTR5 A0A182M4W7 A0A158NTI8 E0VEZ5 B0WAT2 A0A182QCP7 A0A182WEI4 A0A151X3D8 A0A195EG06 A0A0M9A0M7 A0A151INR9 A0A336LNL9 A0A0P5U5C6 Q7PZ11 A0A0P5CSM7 A0A0P5U041 A0A182XKM2 A0A182V284 A0A182L4C0 A0A0P5XL53 A0A182TJL4 A0A182HGX8 A0A0N8A234 A0A182IVK7 E9FZR3 A0A0P6INE2 A0A1B6H3R1 A0A2M4AN40 W5JUP4 A0A3L8DJ74 A0A0P4XTX4 A0A0P5DHX6 A0A182FUT0 A0A195BRP9 A0A195ESN7 D6WNI9 T1HS24 A0A0C9RW60 A0A1Y1MMD9 A0A164UJL5 A0A1J1J8S1 A0A1B0DNX3 A0A0N8AC08 A0A026W853 A0A0L7QUA4 A0A3G2KX35 N6U9K1 A0A2J7PLN3 A0A154P2V5 A0A2L2YKU4 A0A3Q0JLC4 U4UGQ1 A0A2R5LLY4 A0A0A9YNJ7 B7QDX5 A0A131Y1H0 A0A131YK82 L7MLW4 A0A224Z1P1 A0A1I8QB31 A0A0P4W488 A0A1A9W6F2 A0A1A9ZHY3 A0A1B0BZC7
PDB
6F3T
E-value=2.41433e-165,
Score=1496
Ontologies
GO
PANTHER
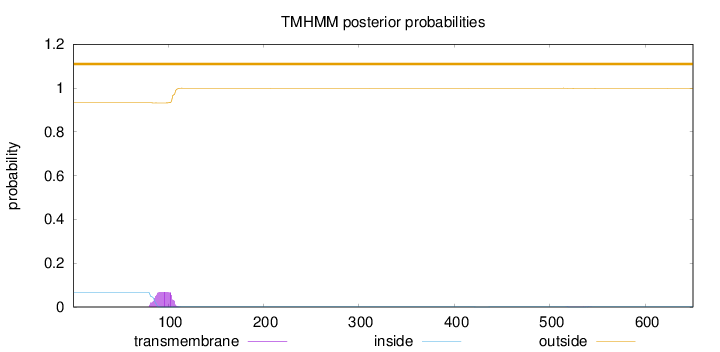
Topology
Length:
650
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.4431
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.06761
outside
1 - 650
Population Genetic Test Statistics
Pi
242.770459
Theta
187.323613
Tajima's D
0.87117
CLR
0.208409
CSRT
0.622068896555172
Interpretation
Uncertain
