Gene
KWMTBOMO01974
Annotation
endonuclease-reverse_transcriptase_[Danaus_plexippus]
Location in the cell
Nuclear Reliability : 4.708
Sequence
CDS
ATGGAAGAAGTTTTAAACACGCTAATTTCAATAAAAAAAGAACTGGAAGAACAAAGAATTGAAATTAGAGAAACTGGAAAAAATGTTACGGAAAAAGTTACACAAAATATTACTAGTATATTCGAGGAGAAATTTTTGGTCTGGGAAGGAAAACTTGGAGAAATGAAAGAAAAAGTGGAAAATCAAGAAAAAAGAATTCATTACCTGGAAAAACAAACAAGAGTAAGAAACTTAGTATTTTTCGGTATCGAGGAAAACGAAACATCATACAAGAGTTTCGAAGAAAATATAATTAAATGGATTGAGCAATATTTCTCTATCAAACTAACCTACAGCGACATTCAAGAGATAAAAAGACTGGGAAAGAAACAAGAGAGACCACGTCCAACTGTTGTAACTTTCACTACACTAGGAATAAAAATCGAAATATTTAAACAAAAAAGAGCGCTGAAAGATACACAGTACTATATGAAAGAAGACTTCCCCAAATATGTACTTGAAAAAAGAAAGGCACTACAGGAACAACTGAAATTAGAGAGAGAAAAGGGAAATACAGCTATACTCAAATATGATAAACTAATTGTATCAAAATACCAAAGTAAAAGAAAATACCGGTTATCTCCAGACAATACCCCACAAATCAATATAGACAAGTATACTCAGGCTAGTAAAAAAAACAAGACAAAGCAGATTGGCAAGACAGTCACTAGGTCAAATAGTATCACAGAAGGAGTCATAAAACCTAGCATGCTCAACTTCTTAGTTAACAAAAATACAATCAAAACTAACAACAACCAAGAAAGTAACAGCGAGAATATATAG
Protein
MEEVLNTLISIKKELEEQRIEIRETGKNVTEKVTQNITSIFEEKFLVWEGKLGEMKEKVENQEKRIHYLEKQTRVRNLVFFGIEENETSYKSFEENIIKWIEQYFSIKLTYSDIQEIKRLGKKQERPRPTVVTFTTLGIKIEIFKQKRALKDTQYYMKEDFPKYVLEKRKALQEQLKLEREKGNTAILKYDKLIVSKYQSKRKYRLSPDNTPQINIDKYTQASKKNKTKQIGKTVTRSNSITEGVIKPSMLNFLVNKNTIKTNNNQESNSENI
Summary
Uniprot
A0A2A4KAH5
A0A0L7KL25
A0A0L7KMI6
A0A194RRA2
D7F178
A0A2H1WF27
+ More
A0A0L7L607 A0A0L7LJT1 A0A0L7KFW9 A0A0L7LLD6 A0A2H1WS99 A0A0L7K2J4 A0A2A4JB54 A0A0L7LIY0 A0A0L7LL17 D7F177 A0A0L7LIU1 A0A0L7LU16 A0A0L7LUV7 A0A0L7L5X6 A0A0L7KKY9 A0A0L7KQ23 A0A2A4J1F3 A0A2A4JED5 A0A0L7LDK8 A0A069DR53 A0A2W1BGL1 A0A2A4J3C0 A0A2A4IX08 A0A2A4K1M2 A0A0L7L3X8 A0A0L7KX87 A0A0L7LNS8 A0A2H1V457 A0A2H1V475 D7F179 A0A0L7L7G4 A0A2A4J942 A0A2A4JJB3 A0A2A4J436 H9J4J3 A0A1Y1KNT7 A0A1S3DLA6 A0A0L7KQ18 A0A1S4EPY6 H9JKN5
A0A0L7L607 A0A0L7LJT1 A0A0L7KFW9 A0A0L7LLD6 A0A2H1WS99 A0A0L7K2J4 A0A2A4JB54 A0A0L7LIY0 A0A0L7LL17 D7F177 A0A0L7LIU1 A0A0L7LU16 A0A0L7LUV7 A0A0L7L5X6 A0A0L7KKY9 A0A0L7KQ23 A0A2A4J1F3 A0A2A4JED5 A0A0L7LDK8 A0A069DR53 A0A2W1BGL1 A0A2A4J3C0 A0A2A4IX08 A0A2A4K1M2 A0A0L7L3X8 A0A0L7KX87 A0A0L7LNS8 A0A2H1V457 A0A2H1V475 D7F179 A0A0L7L7G4 A0A2A4J942 A0A2A4JJB3 A0A2A4J436 H9J4J3 A0A1Y1KNT7 A0A1S3DLA6 A0A0L7KQ18 A0A1S4EPY6 H9JKN5
EMBL
NWSH01000023
PCG80652.1
JTDY01009375
KOB63594.1
JTDY01008567
KOB64512.1
+ More
KQ459995 KPJ18566.1 FJ265563 ADI61831.1 ODYU01008249 SOQ51673.1 JTDY01002701 KOB70892.1 JTDY01000843 KOB75692.1 JTDY01009971 KOB62006.1 JTDY01000640 KOB76373.1 ODYU01010663 SOQ55918.1 JTDY01016948 KOB51874.1 NWSH01002039 PCG69337.1 JTDY01000895 KOB75518.1 JTDY01000780 JTDY01000104 JTDY01000008 KOB75906.1 KOB78842.1 KOB79479.1 FJ265562 ADI61830.1 JTDY01001015 KOB75121.1 JTDY01000100 KOB78869.1 JTDY01000076 KOB79001.1 JTDY01002685 KOB70933.1 JTDY01009161 KOB64038.1 JTDY01007191 KOB65392.1 NWSH01004020 PCG65566.1 NWSH01001771 PCG70189.1 JTDY01001575 KOB73484.1 GBGD01002698 JAC86191.1 KZ150150 PZC72845.1 NWSH01003397 PCG66369.1 NWSH01005231 PCG64345.1 NWSH01000260 PCG77926.1 JTDY01003098 KOB70157.1 JTDY01004672 KOB67888.1 JTDY01000437 KOB77173.1 ODYU01000585 SOQ35630.1 ODYU01000586 SOQ35631.1 FJ265564 ADI61832.1 JTDY01002536 KOB71226.1 NWSH01002532 PCG68054.1 NWSH01001196 PCG72157.1 NWSH01003169 PCG66835.1 BABH01021795 GEZM01082533 JAV61335.1 JTDY01007450 KOB65190.1 BABH01024878
KQ459995 KPJ18566.1 FJ265563 ADI61831.1 ODYU01008249 SOQ51673.1 JTDY01002701 KOB70892.1 JTDY01000843 KOB75692.1 JTDY01009971 KOB62006.1 JTDY01000640 KOB76373.1 ODYU01010663 SOQ55918.1 JTDY01016948 KOB51874.1 NWSH01002039 PCG69337.1 JTDY01000895 KOB75518.1 JTDY01000780 JTDY01000104 JTDY01000008 KOB75906.1 KOB78842.1 KOB79479.1 FJ265562 ADI61830.1 JTDY01001015 KOB75121.1 JTDY01000100 KOB78869.1 JTDY01000076 KOB79001.1 JTDY01002685 KOB70933.1 JTDY01009161 KOB64038.1 JTDY01007191 KOB65392.1 NWSH01004020 PCG65566.1 NWSH01001771 PCG70189.1 JTDY01001575 KOB73484.1 GBGD01002698 JAC86191.1 KZ150150 PZC72845.1 NWSH01003397 PCG66369.1 NWSH01005231 PCG64345.1 NWSH01000260 PCG77926.1 JTDY01003098 KOB70157.1 JTDY01004672 KOB67888.1 JTDY01000437 KOB77173.1 ODYU01000585 SOQ35630.1 ODYU01000586 SOQ35631.1 FJ265564 ADI61832.1 JTDY01002536 KOB71226.1 NWSH01002532 PCG68054.1 NWSH01001196 PCG72157.1 NWSH01003169 PCG66835.1 BABH01021795 GEZM01082533 JAV61335.1 JTDY01007450 KOB65190.1 BABH01024878
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
A0A2A4KAH5
A0A0L7KL25
A0A0L7KMI6
A0A194RRA2
D7F178
A0A2H1WF27
+ More
A0A0L7L607 A0A0L7LJT1 A0A0L7KFW9 A0A0L7LLD6 A0A2H1WS99 A0A0L7K2J4 A0A2A4JB54 A0A0L7LIY0 A0A0L7LL17 D7F177 A0A0L7LIU1 A0A0L7LU16 A0A0L7LUV7 A0A0L7L5X6 A0A0L7KKY9 A0A0L7KQ23 A0A2A4J1F3 A0A2A4JED5 A0A0L7LDK8 A0A069DR53 A0A2W1BGL1 A0A2A4J3C0 A0A2A4IX08 A0A2A4K1M2 A0A0L7L3X8 A0A0L7KX87 A0A0L7LNS8 A0A2H1V457 A0A2H1V475 D7F179 A0A0L7L7G4 A0A2A4J942 A0A2A4JJB3 A0A2A4J436 H9J4J3 A0A1Y1KNT7 A0A1S3DLA6 A0A0L7KQ18 A0A1S4EPY6 H9JKN5
A0A0L7L607 A0A0L7LJT1 A0A0L7KFW9 A0A0L7LLD6 A0A2H1WS99 A0A0L7K2J4 A0A2A4JB54 A0A0L7LIY0 A0A0L7LL17 D7F177 A0A0L7LIU1 A0A0L7LU16 A0A0L7LUV7 A0A0L7L5X6 A0A0L7KKY9 A0A0L7KQ23 A0A2A4J1F3 A0A2A4JED5 A0A0L7LDK8 A0A069DR53 A0A2W1BGL1 A0A2A4J3C0 A0A2A4IX08 A0A2A4K1M2 A0A0L7L3X8 A0A0L7KX87 A0A0L7LNS8 A0A2H1V457 A0A2H1V475 D7F179 A0A0L7L7G4 A0A2A4J942 A0A2A4JJB3 A0A2A4J436 H9J4J3 A0A1Y1KNT7 A0A1S3DLA6 A0A0L7KQ18 A0A1S4EPY6 H9JKN5
Ontologies
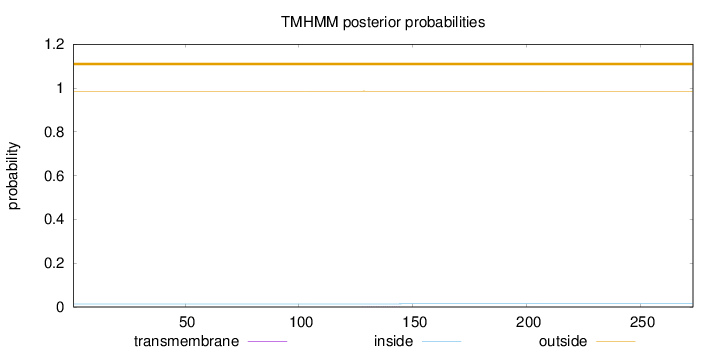
Topology
Length:
273
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00393
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01418
outside
1 - 273
Population Genetic Test Statistics
Pi
249.574007
Theta
178.125664
Tajima's D
1.261956
CLR
134.433335
CSRT
0.726213689315534
Interpretation
Uncertain
