Gene
KWMTBOMO01967
Pre Gene Modal
BGIBMGA014038
Annotation
PREDICTED:_uncharacterized_protein_LOC105841846_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.252 PlasmaMembrane Reliability : 4.922
Sequence
CDS
ATGCAAAACGATAATGGATCATATGGCAGTGCTAGCGATTTGCTGGATAGCCATGTACTAGGAAACGACAAATGTTCTGAACCAGCGATGGAGAGATCTACTAGATCGAGTCTTGATGCCAACAGTGTTGATATCGGGGAAATAACTGAGTCGCCTGATGCCGAGTACATGAGCACCTCAGCAATTTTACATTACTGTAAATCCAAAATTCTGATCCCATATTTAAAATTACTGACAATCATGGGCTTACGACCAGTAATGGACGTCTCTAACACGTCAAGATGCATGATTTGTTGTTCACACTTTCATTCCGCCCAGGTGGCAATTCTTATGCTCTTAGGATATATTTTACAATATATGGCATGCTTTAGGAGAGATAGAGGATTTTGCTATAAATTAGTGCCGTTAGCATTAACTTCGTCTTTAGAATATGACACATATAAAGAAGTATGCTACGGGAATGTTGCTTTTATTTACATAGGACCCAGCGTTTTACATTTTTTGGGATTCTTATATGCACTGTATTTATTTCGGGTTTCCGATAACGAACAGTTACAGAATTTGATGGAAAGAGTTTTTCTTCTATCTTCGTATGCGCCTCGAGGAATGCCAACTGCAAAGCCTAGACGACTGTTGCGTATGTTGTGGTTCTTTATAATTCTTAGTGTGGTTTGGATGTGCTTATCACTTTGTTTGGTGAATATGATGTTGGCCAAAGGTAACATAATATTCAGATGGATGGAAACCAGTTCCTATGAAGTAAACGTGGCTCTGAAGATACTGATGATAATTTGCACACTGATACACGACATGGTACAGGCGACCATAATAACTAGTTACTGTCTCCAAGCACAACTACTCCAAGCTCACCTCATGTTTTTAAGAGAACGTCTGCTCAATAGGACTATGACTCCACTCAATTGGATGAGGGAAATAGCAGAATTCCGTAAATTATTGAAGTATTTGAATGACGATCTAGCGCCAGCGATATGTTTATTCACGATGGTGAACATCTCTTGGGCCATATCAGGAACTATGTGGTTACTGAATTTGGATAAAGTGGACACCGAGACGGAACCAATAATTGGAATTAGCGTTCTTAATCAACTGTTGTGGATATCGGCAGCGGTCGTACCCTTTATTCAGGCGGCCCGGTTGTCTAGAGAGTGCGCTAAGACTCAGTCCGTCGGGCACGAGCTGTGCGTCCGACCGTTTTTACACCAAGACACGTCACAGGAAGACATTACCACGATAATGCTGTACGCGTCGACGCTGCAACTGCACGCCAAACTGTTCCACTACCCTATAGCCGGACGTTACCTCTATCTCGCCTGA
Protein
MQNDNGSYGSASDLLDSHVLGNDKCSEPAMERSTRSSLDANSVDIGEITESPDAEYMSTSAILHYCKSKILIPYLKLLTIMGLRPVMDVSNTSRCMICCSHFHSAQVAILMLLGYILQYMACFRRDRGFCYKLVPLALTSSLEYDTYKEVCYGNVAFIYIGPSVLHFLGFLYALYLFRVSDNEQLQNLMERVFLLSSYAPRGMPTAKPRRLLRMLWFFIILSVVWMCLSLCLVNMMLAKGNIIFRWMETSSYEVNVALKILMIICTLIHDMVQATIITSYCLQAQLLQAHLMFLRERLLNRTMTPLNWMREIAEFRKLLKYLNDDLAPAICLFTMVNISWAISGTMWLLNLDKVDTETEPIIGISVLNQLLWISAAVVPFIQAARLSRECAKTQSVGHELCVRPFLHQDTSQEDITTIMLYASTLQLHAKLFHYPIAGRYLYLA
Summary
Uniprot
A0A2A4JQQ4
A0A2W1BUQ5
A0A194PRR1
A0A194RCZ2
A0A1E1WUR3
A0A212FH71
+ More
A0A0L7LVN4 A0A0L7RA54 E2B8U3 A0A310SGS1 A0A154PRP6 A0A2A3END2 A0A087ZP41 A0A3L8DR78 A0A026WB34 A0A158NHK8 A0A2J7RB08 E2AQ01 K7IQ03 A0A232F1I7 A0A195E2Z8 A0A151WGI3 A0A195C2B6 A0A195AUV9 A0A1Y1N4P4 A0A195ETW2 D6WM94 A0A0M9A2M6 A0A1W4XUN2 N6SYP9 F4WA63 U4UQT5 A0A0J7KWI0 E0VBH1 A0A0T6AVH1 A0A084WAV1 A0A182JMN9 A0A1I8MWH1 A0A1I8MWG4 A0A336KLD4 Q7Q8V7 A0A1B0CRZ0 B4MNM2 A0A0Q9W314 A0A182MYB0 A0A1W4VR52 B4LK24 A0A1W4VDP1 A0A1I8JT77 B3MEM2 A0A182Q439 A0A0A1WUY8 A0A0Q9XJK4 A0A0K8VM99 B4KM19 C0MJ87 B4HBF2 A0A0B4KFG3 A0ANU3 C0MJ80 C0MJ84 Q9W1W8 A0ANU8 W8BRR4 N6W6U2 B4P908 Q28WZ7 B4J4R4 B3NNZ2 A0A0J9RJC4 A0A0M3QV69 B4I8E3 A0A0J9U8Z6 A0A3B0IZ04 A0A1I8NM86 A0A1D2NA85 A0A182RZ15 W5JGI5 B4QI37 A0A182JUH9 A0A0P6IN82 A0A0P5XY35 A0A0P6CHC2 A0A0P4XIB0 A0A0P4Y782 A0A293L9P6 A0A1A9WW19 A0A1S3JN93 A0A0P5TG88
A0A0L7LVN4 A0A0L7RA54 E2B8U3 A0A310SGS1 A0A154PRP6 A0A2A3END2 A0A087ZP41 A0A3L8DR78 A0A026WB34 A0A158NHK8 A0A2J7RB08 E2AQ01 K7IQ03 A0A232F1I7 A0A195E2Z8 A0A151WGI3 A0A195C2B6 A0A195AUV9 A0A1Y1N4P4 A0A195ETW2 D6WM94 A0A0M9A2M6 A0A1W4XUN2 N6SYP9 F4WA63 U4UQT5 A0A0J7KWI0 E0VBH1 A0A0T6AVH1 A0A084WAV1 A0A182JMN9 A0A1I8MWH1 A0A1I8MWG4 A0A336KLD4 Q7Q8V7 A0A1B0CRZ0 B4MNM2 A0A0Q9W314 A0A182MYB0 A0A1W4VR52 B4LK24 A0A1W4VDP1 A0A1I8JT77 B3MEM2 A0A182Q439 A0A0A1WUY8 A0A0Q9XJK4 A0A0K8VM99 B4KM19 C0MJ87 B4HBF2 A0A0B4KFG3 A0ANU3 C0MJ80 C0MJ84 Q9W1W8 A0ANU8 W8BRR4 N6W6U2 B4P908 Q28WZ7 B4J4R4 B3NNZ2 A0A0J9RJC4 A0A0M3QV69 B4I8E3 A0A0J9U8Z6 A0A3B0IZ04 A0A1I8NM86 A0A1D2NA85 A0A182RZ15 W5JGI5 B4QI37 A0A182JUH9 A0A0P6IN82 A0A0P5XY35 A0A0P6CHC2 A0A0P4XIB0 A0A0P4Y782 A0A293L9P6 A0A1A9WW19 A0A1S3JN93 A0A0P5TG88
Pubmed
28756777
26354079
22118469
26227816
20798317
30249741
+ More
24508170 21347285 20075255 28648823 28004739 18362917 19820115 23537049 21719571 20566863 24438588 25315136 12364791 17994087 25830018 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 16951084 26109357 26109356 24495485 15632085 23185243 17550304 22936249 27289101 20920257 23761445
24508170 21347285 20075255 28648823 28004739 18362917 19820115 23537049 21719571 20566863 24438588 25315136 12364791 17994087 25830018 19126864 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 16951084 26109357 26109356 24495485 15632085 23185243 17550304 22936249 27289101 20920257 23761445
EMBL
NWSH01000880
PCG73740.1
KZ149895
PZC78778.1
KQ459601
KPI93825.1
+ More
KQ460398 KPJ15140.1 GDQN01000488 JAT90566.1 AGBW02008527 OWR53079.1 JTDY01000001 KOB79537.1 KQ414619 KOC67762.1 GL446384 EFN87883.1 KQ761343 OAD57877.1 KQ435090 KZC14595.1 KZ288204 PBC33313.1 QOIP01000005 RLU22945.1 KK107293 EZA53312.1 ADTU01015916 NEVH01006564 PNF38019.1 GL441645 EFN64488.1 NNAY01001260 OXU24581.1 KQ979763 KYN19287.1 KQ983167 KYQ46949.1 KQ978350 KYM94730.1 KQ976737 KYM75961.1 GEZM01012954 JAV92852.1 KQ981975 KYN31596.1 KQ971343 EFA04239.2 KQ435784 KOX74559.1 APGK01051866 KB741207 ENN72904.1 GL888041 EGI68919.1 KB632327 ERL92521.1 LBMM01002572 KMQ94643.1 DS235030 EEB10727.1 LJIG01022707 KRT79132.1 ATLV01022269 KE525331 KFB47345.1 UFQS01000542 UFQT01000542 UFQT01001948 SSX04738.1 SSX25101.1 SSX32215.1 AAAB01008933 EAA09975.4 AJWK01025507 AJWK01025508 CH963848 EDW73711.1 CH940648 KRF79513.1 EDW60613.2 APCN01005582 APCN01005583 CH902619 EDV37642.2 AXCN02002043 AXCN02002044 AXCN02002045 GBXI01012049 JAD02243.1 CH933808 KRG05492.1 GDHF01012336 JAI39978.1 EDW10808.2 FM245409 CAR93335.1 CH479252 EDW38694.1 AE013599 AGB93677.1 AM294331 CAL26261.1 FM245402 CAR93328.1 FM245406 CAR93332.1 AY069282 AM294332 AM294333 AM294334 AM294335 AM294337 AM294338 AM294339 FM245399 FM245400 FM245401 FM245403 FM245404 FM245405 FM245407 FM245408 AAF46934.2 AAL39427.1 CAL26262.1 CAL26263.1 CAL26264.1 CAL26265.1 CAL26267.1 CAL26268.1 CAL26269.1 CAR93325.1 CAR93326.1 CAR93327.1 CAR93329.1 CAR93330.1 CAR93331.1 CAR93333.1 CAR93334.1 AM294336 CAL26266.1 GAMC01002535 JAC04021.1 CM000071 ENO01953.2 KRT03153.1 CM000158 EDW92248.1 EAL26519.2 CH916367 EDW00610.1 CH954179 EDV56725.1 CM002911 KMY95936.1 CP012524 ALC41863.1 CH480824 EDW56868.1 KMY95935.1 OUUW01000001 SPP73295.1 LJIJ01000130 ODN01995.1 ADMH02001479 ETN62428.1 CM000362 EDX08291.1 GDIQ01255719 GDIQ01029077 JAN65660.1 GDIP01065557 LRGB01003257 JAM38158.1 KZS03477.1 GDIP01015168 JAM88547.1 GDIP01241426 JAI81975.1 GDIP01232582 JAI90819.1 GFWV01005217 MAA29947.1 GDIP01126650 JAL77064.1
KQ460398 KPJ15140.1 GDQN01000488 JAT90566.1 AGBW02008527 OWR53079.1 JTDY01000001 KOB79537.1 KQ414619 KOC67762.1 GL446384 EFN87883.1 KQ761343 OAD57877.1 KQ435090 KZC14595.1 KZ288204 PBC33313.1 QOIP01000005 RLU22945.1 KK107293 EZA53312.1 ADTU01015916 NEVH01006564 PNF38019.1 GL441645 EFN64488.1 NNAY01001260 OXU24581.1 KQ979763 KYN19287.1 KQ983167 KYQ46949.1 KQ978350 KYM94730.1 KQ976737 KYM75961.1 GEZM01012954 JAV92852.1 KQ981975 KYN31596.1 KQ971343 EFA04239.2 KQ435784 KOX74559.1 APGK01051866 KB741207 ENN72904.1 GL888041 EGI68919.1 KB632327 ERL92521.1 LBMM01002572 KMQ94643.1 DS235030 EEB10727.1 LJIG01022707 KRT79132.1 ATLV01022269 KE525331 KFB47345.1 UFQS01000542 UFQT01000542 UFQT01001948 SSX04738.1 SSX25101.1 SSX32215.1 AAAB01008933 EAA09975.4 AJWK01025507 AJWK01025508 CH963848 EDW73711.1 CH940648 KRF79513.1 EDW60613.2 APCN01005582 APCN01005583 CH902619 EDV37642.2 AXCN02002043 AXCN02002044 AXCN02002045 GBXI01012049 JAD02243.1 CH933808 KRG05492.1 GDHF01012336 JAI39978.1 EDW10808.2 FM245409 CAR93335.1 CH479252 EDW38694.1 AE013599 AGB93677.1 AM294331 CAL26261.1 FM245402 CAR93328.1 FM245406 CAR93332.1 AY069282 AM294332 AM294333 AM294334 AM294335 AM294337 AM294338 AM294339 FM245399 FM245400 FM245401 FM245403 FM245404 FM245405 FM245407 FM245408 AAF46934.2 AAL39427.1 CAL26262.1 CAL26263.1 CAL26264.1 CAL26265.1 CAL26267.1 CAL26268.1 CAL26269.1 CAR93325.1 CAR93326.1 CAR93327.1 CAR93329.1 CAR93330.1 CAR93331.1 CAR93333.1 CAR93334.1 AM294336 CAL26266.1 GAMC01002535 JAC04021.1 CM000071 ENO01953.2 KRT03153.1 CM000158 EDW92248.1 EAL26519.2 CH916367 EDW00610.1 CH954179 EDV56725.1 CM002911 KMY95936.1 CP012524 ALC41863.1 CH480824 EDW56868.1 KMY95935.1 OUUW01000001 SPP73295.1 LJIJ01000130 ODN01995.1 ADMH02001479 ETN62428.1 CM000362 EDX08291.1 GDIQ01255719 GDIQ01029077 JAN65660.1 GDIP01065557 LRGB01003257 JAM38158.1 KZS03477.1 GDIP01015168 JAM88547.1 GDIP01241426 JAI81975.1 GDIP01232582 JAI90819.1 GFWV01005217 MAA29947.1 GDIP01126650 JAL77064.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
UP000053825
+ More
UP000008237 UP000076502 UP000242457 UP000005203 UP000279307 UP000053097 UP000005205 UP000235965 UP000000311 UP000002358 UP000215335 UP000078492 UP000075809 UP000078542 UP000078540 UP000078541 UP000007266 UP000053105 UP000192223 UP000019118 UP000007755 UP000030742 UP000036403 UP000009046 UP000030765 UP000075880 UP000095301 UP000007062 UP000092461 UP000007798 UP000008792 UP000075884 UP000192221 UP000075840 UP000007801 UP000075886 UP000009192 UP000008744 UP000000803 UP000001819 UP000002282 UP000001070 UP000008711 UP000092553 UP000001292 UP000268350 UP000095300 UP000094527 UP000075900 UP000000673 UP000000304 UP000075881 UP000076858 UP000091820 UP000085678
UP000008237 UP000076502 UP000242457 UP000005203 UP000279307 UP000053097 UP000005205 UP000235965 UP000000311 UP000002358 UP000215335 UP000078492 UP000075809 UP000078542 UP000078540 UP000078541 UP000007266 UP000053105 UP000192223 UP000019118 UP000007755 UP000030742 UP000036403 UP000009046 UP000030765 UP000075880 UP000095301 UP000007062 UP000092461 UP000007798 UP000008792 UP000075884 UP000192221 UP000075840 UP000007801 UP000075886 UP000009192 UP000008744 UP000000803 UP000001819 UP000002282 UP000001070 UP000008711 UP000092553 UP000001292 UP000268350 UP000095300 UP000094527 UP000075900 UP000000673 UP000000304 UP000075881 UP000076858 UP000091820 UP000085678
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JQQ4
A0A2W1BUQ5
A0A194PRR1
A0A194RCZ2
A0A1E1WUR3
A0A212FH71
+ More
A0A0L7LVN4 A0A0L7RA54 E2B8U3 A0A310SGS1 A0A154PRP6 A0A2A3END2 A0A087ZP41 A0A3L8DR78 A0A026WB34 A0A158NHK8 A0A2J7RB08 E2AQ01 K7IQ03 A0A232F1I7 A0A195E2Z8 A0A151WGI3 A0A195C2B6 A0A195AUV9 A0A1Y1N4P4 A0A195ETW2 D6WM94 A0A0M9A2M6 A0A1W4XUN2 N6SYP9 F4WA63 U4UQT5 A0A0J7KWI0 E0VBH1 A0A0T6AVH1 A0A084WAV1 A0A182JMN9 A0A1I8MWH1 A0A1I8MWG4 A0A336KLD4 Q7Q8V7 A0A1B0CRZ0 B4MNM2 A0A0Q9W314 A0A182MYB0 A0A1W4VR52 B4LK24 A0A1W4VDP1 A0A1I8JT77 B3MEM2 A0A182Q439 A0A0A1WUY8 A0A0Q9XJK4 A0A0K8VM99 B4KM19 C0MJ87 B4HBF2 A0A0B4KFG3 A0ANU3 C0MJ80 C0MJ84 Q9W1W8 A0ANU8 W8BRR4 N6W6U2 B4P908 Q28WZ7 B4J4R4 B3NNZ2 A0A0J9RJC4 A0A0M3QV69 B4I8E3 A0A0J9U8Z6 A0A3B0IZ04 A0A1I8NM86 A0A1D2NA85 A0A182RZ15 W5JGI5 B4QI37 A0A182JUH9 A0A0P6IN82 A0A0P5XY35 A0A0P6CHC2 A0A0P4XIB0 A0A0P4Y782 A0A293L9P6 A0A1A9WW19 A0A1S3JN93 A0A0P5TG88
A0A0L7LVN4 A0A0L7RA54 E2B8U3 A0A310SGS1 A0A154PRP6 A0A2A3END2 A0A087ZP41 A0A3L8DR78 A0A026WB34 A0A158NHK8 A0A2J7RB08 E2AQ01 K7IQ03 A0A232F1I7 A0A195E2Z8 A0A151WGI3 A0A195C2B6 A0A195AUV9 A0A1Y1N4P4 A0A195ETW2 D6WM94 A0A0M9A2M6 A0A1W4XUN2 N6SYP9 F4WA63 U4UQT5 A0A0J7KWI0 E0VBH1 A0A0T6AVH1 A0A084WAV1 A0A182JMN9 A0A1I8MWH1 A0A1I8MWG4 A0A336KLD4 Q7Q8V7 A0A1B0CRZ0 B4MNM2 A0A0Q9W314 A0A182MYB0 A0A1W4VR52 B4LK24 A0A1W4VDP1 A0A1I8JT77 B3MEM2 A0A182Q439 A0A0A1WUY8 A0A0Q9XJK4 A0A0K8VM99 B4KM19 C0MJ87 B4HBF2 A0A0B4KFG3 A0ANU3 C0MJ80 C0MJ84 Q9W1W8 A0ANU8 W8BRR4 N6W6U2 B4P908 Q28WZ7 B4J4R4 B3NNZ2 A0A0J9RJC4 A0A0M3QV69 B4I8E3 A0A0J9U8Z6 A0A3B0IZ04 A0A1I8NM86 A0A1D2NA85 A0A182RZ15 W5JGI5 B4QI37 A0A182JUH9 A0A0P6IN82 A0A0P5XY35 A0A0P6CHC2 A0A0P4XIB0 A0A0P4Y782 A0A293L9P6 A0A1A9WW19 A0A1S3JN93 A0A0P5TG88
Ontologies
PANTHER
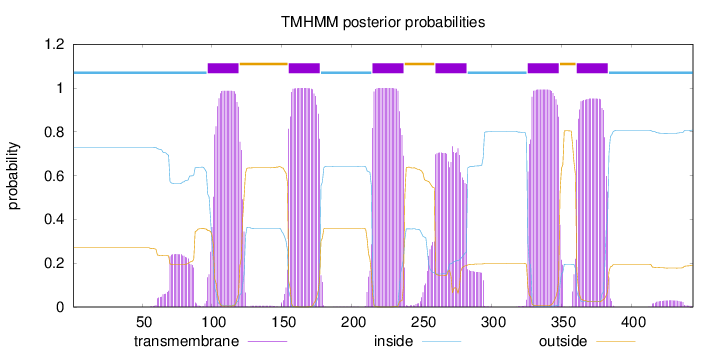
Topology
Length:
444
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
134.87575
Exp number, first 60 AAs:
0.02214
Total prob of N-in:
0.72948
inside
1 - 96
TMhelix
97 - 119
outside
120 - 154
TMhelix
155 - 177
inside
178 - 214
TMhelix
215 - 237
outside
238 - 259
TMhelix
260 - 282
inside
283 - 325
TMhelix
326 - 348
outside
349 - 360
TMhelix
361 - 383
inside
384 - 444
Population Genetic Test Statistics
Pi
251.19031
Theta
186.659691
Tajima's D
1.96068
CLR
0.15769
CSRT
0.875106244687766
Interpretation
Uncertain
