Gene
KWMTBOMO01961
Pre Gene Modal
BGIBMGA001343
Annotation
putative_Aktip_protein_[Operophtera_brumata]
Full name
Protein crossbronx homolog
+ More
AKT-interacting protein
AKT-interacting protein
Alternative Name
Fused toes protein homolog
Ft1
FT1
Fused toes protein
Ft1
FT1
Fused toes protein
Location in the cell
Nuclear Reliability : 2.29
Sequence
CDS
ATGACAGAATACCGTATGTTGCAAACAGAAAATTTACGTGGAATCTATGTTATACCTTCTTATGAGAGTTCATTTCTTTGGTACGGCGTGATTTTTGTTCGATCTGGTCTCTACGAAGGAGGTGTATTTAGATTTACAATTACATTGCCTGAAAAATTTCCAGACGATGAAACACCTGTTTTGACTTTTGTGTCAAACTTATATCATCCTGCTATTGACGCAAGCACTGGCCTTTTAAGTCTGAGAGAAGTGTTTCCAATGTGGGACAAAAAGCGTGACCATGTCTGGCAAATACTCAAATATTTACATATAATATTTGATAATTTAAGCATCAAGGTACCTGCCAATACTGAAGCAGCATGTATGTACAAAACAAATCGAAAATTCTTTATGCAAAAAGTTAGAGAATGCGTTGTTACTAGCATAGATCACATATATGATGATCCACCTACAGAAGACAAACATTATATAACTTTTAAACCTTATGATCCTGATATACATGAAAAAGCAAAGGACATAATGCTTACAGCTTTTAAACCAACTGAAGGAGCCACTCAAGGGATGTCATGGGTTCAACAAGGATGCTACCAACCATTTTCAAAAGAAGAAACAATGTCATAG
Protein
MTEYRMLQTENLRGIYVIPSYESSFLWYGVIFVRSGLYEGGVFRFTITLPEKFPDDETPVLTFVSNLYHPAIDASTGLLSLREVFPMWDKKRDHVWQILKYLHIIFDNLSIKVPANTEAACMYKTNRKFFMQKVRECVVTSIDHIYDDPPTEDKHYITFKPYDPDIHEKAKDIMLTAFKPTEGATQGMSWVQQGCYQPFSKEETMS
Summary
Description
Component of the FTS/Hook/FHIP complex (FHF complex). The FHF complex may function to promote vesicle trafficking and/or fusion via the homotypic vesicular protein sorting complex (the HOPS complex). Regulates apoptosis by enhancing phosphorylation and activation of AKT1. Increases release of TNFSF6 via the AKT1/GSK3B/NFATC1 signaling cascade (By similarity).
Component of the FTS/Hook/FHIP complex (FHF complex). The FHF complex may function to promote vesicle trafficking and/or fusion via the homotypic vesicular protein sorting complex (the HOPS complex). Regulates apoptosis by enhancing phosphorylation and activation of AKT1. Increases release of TNFSF6 via the AKT1/GSK3B/NFATC1 signaling cascade.
Component of the FTS/Hook/FHIP complex (FHF complex). The FHF complex may function to promote vesicle trafficking and/or fusion via the homotypic vesicular protein sorting complex (the HOPS complex). Regulates apoptosis by enhancing phosphorylation and activation of AKT1. Increases release of TNFSF6 via the AKT1/GSK3B/NFATC1 signaling cascade.
Subunit
Component of the FTS/Hook/FHIP complex (FHF complex), composed of AKTIP/FTS, FAM160A2, and one or more members of the Hook family of proteins HOOK1, HOOK2, and HOOK3. May interact directly with HOOK1, HOOK2 and HOOK3. The FHF complex associates with the homotypic vesicular sorting complex (the HOPS complex). Also interacts with AKT1 (By similarity).
Component of the FTS/Hook/FHIP complex (FHF complex), composed of AKTIP/FTS, FAM160A2, and one or more members of the Hook family of proteins HOOK1, HOOK2, and HOOK3. May interact directly with HOOK1, HOOK2 and HOOK3. The FHF complex associates with the homotypic vesicular sorting complex (the HOPS complex). Also interacts with AKT1.
Component of the FTS/Hook/FHIP complex (FHF complex), composed of AKTIP/FTS, FAM160A2, and one or more members of the Hook family of proteins HOOK1, HOOK2, and HOOK3. May interact directly with HOOK1, HOOK2 and HOOK3. The FHF complex associates with the homotypic vesicular sorting complex (the HOPS complex). Also interacts with AKT1.
Similarity
Belongs to the ubiquitin-conjugating enzyme family. FTS subfamily.
Keywords
Complete proteome
Reference proteome
Apoptosis
Cell membrane
Cytoplasm
Membrane
Phosphoprotein
Protein transport
Transport
Alternative splicing
Feature
chain Protein crossbronx homolog
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9IVR2
A0A0L7LWN7
A0A2W1BUM6
A0A1E1WA59
A0A2A4JQ69
A0A2H1WZD7
+ More
A0A194RGU0 A0A194PRR6 A0A212FH65 S4PD56 D6WJD8 A0A1Y1KFX7 A0A182GXK0 A0A2M4AT28 A0A0K8TT76 K7J900 A0A182QB89 A0A2M4BZN6 A0A1I8N4N8 A0A182IGC0 A0A182YIU2 W5JG73 Q7PRH1 A0A2M3ZBI9 A0A2M3ZBD1 A0A182W3X6 W8AZL0 A0A0L0BMG3 Q17PP1 A0A182FBW1 A0A0M9A7F6 A0A2K7P682 A0A182JV24 A0A0K8VIG2 A0A034VM89 A0A084VUQ5 A0A0A1XL19 A0A1I8PW02 N6ULD7 A0A182LND3 A0A182SEI6 B0X6E8 A0A1Q3FIJ7 A0A340XHF7 A0A182RJT4 A0A2G8K6C8 A0A154P381 A0A2Y9SXY8 A0A2Y9LM75 A0A341BN08 V5GTJ9 A0A2U4B4E0 U5EX63 A0A067RLL8 A0A091GK21 A0A0L7QXF7 A0A1S3WVD2 A0A1V4K1V2 J3JT61 A0A2I0M2H7 A0A195CNW0 A0A2Y9QL86 A0A1U7UKX7 G1SIJ3 A0A2Y9ICS3 Q5FVH4 I3MC17 A0A2U3VLL5 A0A2J8VYJ5 A0A2J7QY33 A0A3P4RWG4 A0A2U3XMF9 A0A2K6CL88 G3RFT1 A0A2I3HBY3 A0A0D9QZ26 A0A096NIC1 A0A2K6QIE4 A0A2K6TV23 A0A2J8MWV9 I0FHV9 A0A024R6T5 Q4R5E1 Q9H8T0 A0A1D5QUZ4 A0A2Y9KKW0 M3XW49 U3ENC0 A0A3Q0CBV3 M1EDW4 A0A1U8CRZ6 A0A1W7RJG5 A0A151MS13 E2BC10 A0A1S3G411 A0A182PLT8 A0A3P8VIW0 A0A340XJR4 D3Z2J4 Q64362
A0A194RGU0 A0A194PRR6 A0A212FH65 S4PD56 D6WJD8 A0A1Y1KFX7 A0A182GXK0 A0A2M4AT28 A0A0K8TT76 K7J900 A0A182QB89 A0A2M4BZN6 A0A1I8N4N8 A0A182IGC0 A0A182YIU2 W5JG73 Q7PRH1 A0A2M3ZBI9 A0A2M3ZBD1 A0A182W3X6 W8AZL0 A0A0L0BMG3 Q17PP1 A0A182FBW1 A0A0M9A7F6 A0A2K7P682 A0A182JV24 A0A0K8VIG2 A0A034VM89 A0A084VUQ5 A0A0A1XL19 A0A1I8PW02 N6ULD7 A0A182LND3 A0A182SEI6 B0X6E8 A0A1Q3FIJ7 A0A340XHF7 A0A182RJT4 A0A2G8K6C8 A0A154P381 A0A2Y9SXY8 A0A2Y9LM75 A0A341BN08 V5GTJ9 A0A2U4B4E0 U5EX63 A0A067RLL8 A0A091GK21 A0A0L7QXF7 A0A1S3WVD2 A0A1V4K1V2 J3JT61 A0A2I0M2H7 A0A195CNW0 A0A2Y9QL86 A0A1U7UKX7 G1SIJ3 A0A2Y9ICS3 Q5FVH4 I3MC17 A0A2U3VLL5 A0A2J8VYJ5 A0A2J7QY33 A0A3P4RWG4 A0A2U3XMF9 A0A2K6CL88 G3RFT1 A0A2I3HBY3 A0A0D9QZ26 A0A096NIC1 A0A2K6QIE4 A0A2K6TV23 A0A2J8MWV9 I0FHV9 A0A024R6T5 Q4R5E1 Q9H8T0 A0A1D5QUZ4 A0A2Y9KKW0 M3XW49 U3ENC0 A0A3Q0CBV3 M1EDW4 A0A1U8CRZ6 A0A1W7RJG5 A0A151MS13 E2BC10 A0A1S3G411 A0A182PLT8 A0A3P8VIW0 A0A340XJR4 D3Z2J4 Q64362
Pubmed
19121390
26227816
28756777
26354079
22118469
23622113
+ More
18362917 19820115 28004739 26483478 26369729 20075255 25315136 25244985 20920257 23761445 12364791 24495485 26108605 17510324 25348373 24438588 25830018 23537049 20966253 29023486 24845553 22516182 23371554 21993624 15489334 22673903 25362486 16136131 25319552 11181995 14702039 14749367 18799622 23186163 17431167 25243066 28071753 23236062 26358130 22293439 20798317 24487278 19468303 21183079 9383278 16141072 7956835
18362917 19820115 28004739 26483478 26369729 20075255 25315136 25244985 20920257 23761445 12364791 24495485 26108605 17510324 25348373 24438588 25830018 23537049 20966253 29023486 24845553 22516182 23371554 21993624 15489334 22673903 25362486 16136131 25319552 11181995 14702039 14749367 18799622 23186163 17431167 25243066 28071753 23236062 26358130 22293439 20798317 24487278 19468303 21183079 9383278 16141072 7956835
EMBL
BABH01042430
JTDY01000001
KOB79536.1
KZ149895
PZC78782.1
GDQN01007164
+ More
JAT83890.1 NWSH01000880 PCG73744.1 ODYU01012251 SOQ58460.1 KQ460398 KPJ15146.1 KQ459601 KPI93830.1 AGBW02008527 OWR53086.1 GAIX01003796 JAA88764.1 KQ971343 EFA04459.2 GEZM01084914 GEZM01084912 JAV60409.1 JXUM01020608 JXUM01020609 JXUM01020610 JXUM01020611 KQ560576 KXJ81839.1 GGFK01010634 MBW43955.1 GDAI01000493 JAI17110.1 AXCN02002125 GGFJ01009057 MBW58198.1 APCN01005189 ADMH02001609 ETN61814.1 AAAB01008849 GGFM01005121 MBW25872.1 GGFM01005007 MBW25758.1 GAMC01012280 JAB94275.1 JRES01001654 KNC21123.1 CH477190 KQ435719 KOX78780.1 GDHF01013656 JAI38658.1 GAKP01014501 JAC44451.1 ATLV01016986 KE525139 KFB41699.1 GBXI01011895 GBXI01002650 GBXI01000709 JAD02397.1 JAD11642.1 JAD13583.1 APGK01028100 KB740686 KB632335 ENN79512.1 ERL92817.1 DS232412 GFDL01007635 JAV27410.1 MRZV01000841 PIK43568.1 KQ434809 KZC06376.1 GALX01000982 JAB67484.1 GANO01001103 JAB58768.1 KK852435 KDR23933.1 KL447522 KFO74472.1 KQ414704 KOC63302.1 LSYS01005191 OPJ78361.1 BT126417 AEE61381.1 AKCR02000046 PKK23885.1 KQ977481 KYN02423.1 AAGW02052744 BC089985 AGTP01009331 NDHI03003406 PNJ62546.1 PNJ62549.1 NEVH01009368 PNF33495.1 CYRY02043463 VCX37878.1 CABD030101085 ADFV01012811 AQIB01137407 AHZZ02018736 AACZ04030017 NBAG03000242 PNI63995.1 PNI63997.1 JV043964 JV635819 AFI34035.1 AFJ71159.1 CH471092 EAW82809.1 AB169603 AK023320 CR457308 AK222743 BC001134 BC095401 JSUE03026230 AEYP01015437 GAMS01005121 GAMS01005120 GAMR01006846 GAMR01006845 GAMQ01004716 JAB18015.1 JAB27086.1 JAB37135.1 JP005411 AER94008.1 GDAY02000110 JAV51303.1 AKHW03005226 KYO27324.1 GL447244 EFN86764.1 AC122441 AC139351 Z67963 X71978 AK146459 AK158704 CH466525 BC008130
JAT83890.1 NWSH01000880 PCG73744.1 ODYU01012251 SOQ58460.1 KQ460398 KPJ15146.1 KQ459601 KPI93830.1 AGBW02008527 OWR53086.1 GAIX01003796 JAA88764.1 KQ971343 EFA04459.2 GEZM01084914 GEZM01084912 JAV60409.1 JXUM01020608 JXUM01020609 JXUM01020610 JXUM01020611 KQ560576 KXJ81839.1 GGFK01010634 MBW43955.1 GDAI01000493 JAI17110.1 AXCN02002125 GGFJ01009057 MBW58198.1 APCN01005189 ADMH02001609 ETN61814.1 AAAB01008849 GGFM01005121 MBW25872.1 GGFM01005007 MBW25758.1 GAMC01012280 JAB94275.1 JRES01001654 KNC21123.1 CH477190 KQ435719 KOX78780.1 GDHF01013656 JAI38658.1 GAKP01014501 JAC44451.1 ATLV01016986 KE525139 KFB41699.1 GBXI01011895 GBXI01002650 GBXI01000709 JAD02397.1 JAD11642.1 JAD13583.1 APGK01028100 KB740686 KB632335 ENN79512.1 ERL92817.1 DS232412 GFDL01007635 JAV27410.1 MRZV01000841 PIK43568.1 KQ434809 KZC06376.1 GALX01000982 JAB67484.1 GANO01001103 JAB58768.1 KK852435 KDR23933.1 KL447522 KFO74472.1 KQ414704 KOC63302.1 LSYS01005191 OPJ78361.1 BT126417 AEE61381.1 AKCR02000046 PKK23885.1 KQ977481 KYN02423.1 AAGW02052744 BC089985 AGTP01009331 NDHI03003406 PNJ62546.1 PNJ62549.1 NEVH01009368 PNF33495.1 CYRY02043463 VCX37878.1 CABD030101085 ADFV01012811 AQIB01137407 AHZZ02018736 AACZ04030017 NBAG03000242 PNI63995.1 PNI63997.1 JV043964 JV635819 AFI34035.1 AFJ71159.1 CH471092 EAW82809.1 AB169603 AK023320 CR457308 AK222743 BC001134 BC095401 JSUE03026230 AEYP01015437 GAMS01005121 GAMS01005120 GAMR01006846 GAMR01006845 GAMQ01004716 JAB18015.1 JAB27086.1 JAB37135.1 JP005411 AER94008.1 GDAY02000110 JAV51303.1 AKHW03005226 KYO27324.1 GL447244 EFN86764.1 AC122441 AC139351 Z67963 X71978 AK146459 AK158704 CH466525 BC008130
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000007266 UP000069940 UP000249989 UP000002358 UP000075886 UP000095301 UP000075840 UP000076408 UP000000673 UP000007062 UP000075920 UP000037069 UP000008820 UP000069272 UP000053105 UP000075881 UP000030765 UP000095300 UP000019118 UP000030742 UP000075882 UP000075901 UP000002320 UP000265300 UP000075900 UP000230750 UP000076502 UP000248483 UP000252040 UP000245320 UP000027135 UP000053760 UP000053825 UP000079721 UP000190648 UP000053872 UP000078542 UP000248480 UP000189704 UP000001811 UP000248481 UP000002494 UP000005215 UP000245340 UP000235965 UP000245341 UP000233120 UP000001519 UP000001073 UP000029965 UP000028761 UP000233200 UP000233220 UP000002277 UP000233100 UP000005640 UP000006718 UP000248482 UP000000715 UP000189706 UP000050525 UP000008237 UP000081671 UP000075885 UP000265120 UP000000589
UP000007266 UP000069940 UP000249989 UP000002358 UP000075886 UP000095301 UP000075840 UP000076408 UP000000673 UP000007062 UP000075920 UP000037069 UP000008820 UP000069272 UP000053105 UP000075881 UP000030765 UP000095300 UP000019118 UP000030742 UP000075882 UP000075901 UP000002320 UP000265300 UP000075900 UP000230750 UP000076502 UP000248483 UP000252040 UP000245320 UP000027135 UP000053760 UP000053825 UP000079721 UP000190648 UP000053872 UP000078542 UP000248480 UP000189704 UP000001811 UP000248481 UP000002494 UP000005215 UP000245340 UP000235965 UP000245341 UP000233120 UP000001519 UP000001073 UP000029965 UP000028761 UP000233200 UP000233220 UP000002277 UP000233100 UP000005640 UP000006718 UP000248482 UP000000715 UP000189706 UP000050525 UP000008237 UP000081671 UP000075885 UP000265120 UP000000589
Pfam
PF00179 UQ_con
SUPFAM
SSF54495
SSF54495
Gene 3D
CDD
ProteinModelPortal
H9IVR2
A0A0L7LWN7
A0A2W1BUM6
A0A1E1WA59
A0A2A4JQ69
A0A2H1WZD7
+ More
A0A194RGU0 A0A194PRR6 A0A212FH65 S4PD56 D6WJD8 A0A1Y1KFX7 A0A182GXK0 A0A2M4AT28 A0A0K8TT76 K7J900 A0A182QB89 A0A2M4BZN6 A0A1I8N4N8 A0A182IGC0 A0A182YIU2 W5JG73 Q7PRH1 A0A2M3ZBI9 A0A2M3ZBD1 A0A182W3X6 W8AZL0 A0A0L0BMG3 Q17PP1 A0A182FBW1 A0A0M9A7F6 A0A2K7P682 A0A182JV24 A0A0K8VIG2 A0A034VM89 A0A084VUQ5 A0A0A1XL19 A0A1I8PW02 N6ULD7 A0A182LND3 A0A182SEI6 B0X6E8 A0A1Q3FIJ7 A0A340XHF7 A0A182RJT4 A0A2G8K6C8 A0A154P381 A0A2Y9SXY8 A0A2Y9LM75 A0A341BN08 V5GTJ9 A0A2U4B4E0 U5EX63 A0A067RLL8 A0A091GK21 A0A0L7QXF7 A0A1S3WVD2 A0A1V4K1V2 J3JT61 A0A2I0M2H7 A0A195CNW0 A0A2Y9QL86 A0A1U7UKX7 G1SIJ3 A0A2Y9ICS3 Q5FVH4 I3MC17 A0A2U3VLL5 A0A2J8VYJ5 A0A2J7QY33 A0A3P4RWG4 A0A2U3XMF9 A0A2K6CL88 G3RFT1 A0A2I3HBY3 A0A0D9QZ26 A0A096NIC1 A0A2K6QIE4 A0A2K6TV23 A0A2J8MWV9 I0FHV9 A0A024R6T5 Q4R5E1 Q9H8T0 A0A1D5QUZ4 A0A2Y9KKW0 M3XW49 U3ENC0 A0A3Q0CBV3 M1EDW4 A0A1U8CRZ6 A0A1W7RJG5 A0A151MS13 E2BC10 A0A1S3G411 A0A182PLT8 A0A3P8VIW0 A0A340XJR4 D3Z2J4 Q64362
A0A194RGU0 A0A194PRR6 A0A212FH65 S4PD56 D6WJD8 A0A1Y1KFX7 A0A182GXK0 A0A2M4AT28 A0A0K8TT76 K7J900 A0A182QB89 A0A2M4BZN6 A0A1I8N4N8 A0A182IGC0 A0A182YIU2 W5JG73 Q7PRH1 A0A2M3ZBI9 A0A2M3ZBD1 A0A182W3X6 W8AZL0 A0A0L0BMG3 Q17PP1 A0A182FBW1 A0A0M9A7F6 A0A2K7P682 A0A182JV24 A0A0K8VIG2 A0A034VM89 A0A084VUQ5 A0A0A1XL19 A0A1I8PW02 N6ULD7 A0A182LND3 A0A182SEI6 B0X6E8 A0A1Q3FIJ7 A0A340XHF7 A0A182RJT4 A0A2G8K6C8 A0A154P381 A0A2Y9SXY8 A0A2Y9LM75 A0A341BN08 V5GTJ9 A0A2U4B4E0 U5EX63 A0A067RLL8 A0A091GK21 A0A0L7QXF7 A0A1S3WVD2 A0A1V4K1V2 J3JT61 A0A2I0M2H7 A0A195CNW0 A0A2Y9QL86 A0A1U7UKX7 G1SIJ3 A0A2Y9ICS3 Q5FVH4 I3MC17 A0A2U3VLL5 A0A2J8VYJ5 A0A2J7QY33 A0A3P4RWG4 A0A2U3XMF9 A0A2K6CL88 G3RFT1 A0A2I3HBY3 A0A0D9QZ26 A0A096NIC1 A0A2K6QIE4 A0A2K6TV23 A0A2J8MWV9 I0FHV9 A0A024R6T5 Q4R5E1 Q9H8T0 A0A1D5QUZ4 A0A2Y9KKW0 M3XW49 U3ENC0 A0A3Q0CBV3 M1EDW4 A0A1U8CRZ6 A0A1W7RJG5 A0A151MS13 E2BC10 A0A1S3G411 A0A182PLT8 A0A3P8VIW0 A0A340XJR4 D3Z2J4 Q64362
PDB
6CYO
E-value=1.21851e-10,
Score=156
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Cell membrane
Cell membrane
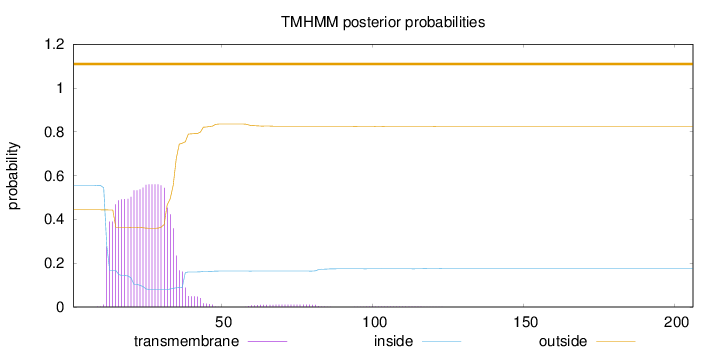
Length:
206
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.5193
Exp number, first 60 AAs:
12.25043
Total prob of N-in:
0.55590
POSSIBLE N-term signal
sequence
outside
1 - 206
Population Genetic Test Statistics
Pi
217.315813
Theta
217.041458
Tajima's D
0.127508
CLR
0.448762
CSRT
0.400629968501575
Interpretation
Uncertain
