Gene
KWMTBOMO01955
Pre Gene Modal
BGIBMGA005921
Annotation
PREDICTED:_EGF_domain-specific_O-linked_N-acetylglucosamine_transferase_[Amyelois_transitella]
Full name
EGF domain-specific O-linked N-acetylglucosamine transferase
Alternative Name
Extracellular O-linked N-acetylglucosamine transferase
Location in the cell
Extracellular Reliability : 1.797 Nuclear Reliability : 1.708
Sequence
CDS
ATGACGAAACAGATTGAATTTTTATGTTTTTTATTATTTCCCATTCACACACTATCAATTAACTTTAATGAATTAAATTTGCCTCCGCAACATATTCCATTCGTTTTTAATGCATTTCCTGAAATTGCTGAAACCTGCAATTCGGAACAAAGCTGCCCTTTCAAGAGTTCGCTGGGTGCAAAATCGTGTTGGGGTTATGAACGAGACTGTGATAAAAAATCCTCGTATCATGTGCGCCCAGTGTGTCCTGGCGATCACAGAGGTTGGGTTAAAACGAAAGAAGCCCAATATGATACGTTTTACACACAAGCAGATTTTGGTTATGTTAAAGAACAAATCGACGATTTGATGGTGATGTGTGAAGCACTATACCCTGATGATACATCGCTTGAATGCTCAAAATATCTTAGGTTCTGCAGGGGAAGAAATATAATGCTGAATTTTACTGGCCTTGTGGGTCGTGGTGATAATTTGAGATATAAAACTGATATTCTCAGTGCTGGACAAATAGGTGGCTACTGTAAGTTCTATTCAGATAGACTTATGAAGGAGGCAGAACATATGAGTGCATTACAGTCATGGGGTCCAGAAATGGTGAATTTCGTCAAAACTCCTAAAAAGCCAATAGCAGATGGAATGTGTGACATTGTCATAGATAAACCAACTTACATCATGAAACTTGATGCTGCTGTTAATATGTATCATCATTTTTGTGACTTTTTCAATTTGTATGCATCGCTACACGTAAATTCCACACATCCATCAACATTTACTAGAAACAATCATATTCTAGTTTGGGAAACATTTACATATGATTCTGCATTCAAAGACGCTTTTAAGGCGTTCACTGAGAATCCGATTTGGGATTTAAAGAGGTTTAGAGGAAAAGTTGTCTGTTTTAAAAATGCAGTGTTCCCGCTTCTGCCAAGAATGATTTTTGGTCTCTACTACAACACACCTCTAATATACGGATGCGAAAGAAGTGGATTGTTTCATGCCTTTTCTAAACACATACTGCATTCTTTGAATATAAAACTGCATATGCGTACCGATGATAGAGTAAGAGTTACTCTCCTATCGAGAGGAACCACATATAGGTCAATTCTAAATGAGAAAGAAATTGTAGATGCTTTGCTCAAAGAAGATGGTTATTATGTCCAACGCGTTGTCTATGACAGGACAGTTTCATTCACAAAGCAGCTAGAAATAACTCACAATACTGATGTGTTCATTGGAATGCATGGAGCCGGTCTGACACATTTACTGTTCTTACCTGATTGGGCAGCGGTATTTGAAATATACAATTGTGAAGATCCTAACTGCTATGCTGATTTAAGTAGACTTCGAGGACTCAAATACGTAACATGGGAAGATAAATCAAAACTTGTCCAACAAGATGAGGGTCATAGTCCCGGTGGTGGTTCACATGCAAAGTTCACCAATTATTCGTTTGACGTCAAAGAATTCTTGAGGTTGGTCGCGAAGTGTGCCGAGTACGTGCGCAACAGACAAGACTTTCAGAACTTCGTTGAAGCGTCTCTTATTAAAATGCACGAAGAATTATAG
Protein
MTKQIEFLCFLLFPIHTLSINFNELNLPPQHIPFVFNAFPEIAETCNSEQSCPFKSSLGAKSCWGYERDCDKKSSYHVRPVCPGDHRGWVKTKEAQYDTFYTQADFGYVKEQIDDLMVMCEALYPDDTSLECSKYLRFCRGRNIMLNFTGLVGRGDNLRYKTDILSAGQIGGYCKFYSDRLMKEAEHMSALQSWGPEMVNFVKTPKKPIADGMCDIVIDKPTYIMKLDAAVNMYHHFCDFFNLYASLHVNSTHPSTFTRNNHILVWETFTYDSAFKDAFKAFTENPIWDLKRFRGKVVCFKNAVFPLLPRMIFGLYYNTPLIYGCERSGLFHAFSKHILHSLNIKLHMRTDDRVRVTLLSRGTTYRSILNEKEIVDALLKEDGYYVQRVVYDRTVSFTKQLEITHNTDVFIGMHGAGLTHLLFLPDWAAVFEIYNCEDPNCYADLSRLRGLKYVTWEDKSKLVQQDEGHSPGGGSHAKFTNYSFDVKEFLRLVAKCAEYVRNRQDFQNFVEASLIKMHEEL
Summary
Description
Catalyzes the transfer of a single N-acetylglucosamine from UDP-GlcNAc to a serine or threonine residue in extracellular proteins resulting in their modification with a beta-linked N-acetylglucosamine (O-GlcNAc). Specifically glycosylates the Thr residue located between the fifth and sixth conserved cysteines of folded EGF-like domains. Involved in epithelial cell adhesion/interaction with the extracellular matrix by mediating glycosylation of proteins in the secretory pathway, such as Dumpy (Dp).
Catalytic Activity
L-seryl-[protein] + UDP-N-acetyl-alpha-D-glucosamine = 3-O-(N-acetyl-beta-D-glucosaminyl)-L-seryl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-glucosamine = 3-O-(N-acetyl-beta-D-glucosaminyl)-L-threonyl-[protein] + H(+) + UDP
L-threonyl-[protein] + UDP-N-acetyl-alpha-D-glucosamine = 3-O-(N-acetyl-beta-D-glucosaminyl)-L-threonyl-[protein] + H(+) + UDP
Cofactor
a divalent metal cation
Keywords
Complete proteome
Endoplasmic reticulum
Glycoprotein
Glycosyltransferase
Reference proteome
Signal
Transferase
Feature
chain EGF domain-specific O-linked N-acetylglucosamine transferase
Uniprot
A0A2A4JSY7
A0A194RGU5
A0A194PRS1
A0A2W1BUZ4
A0A2H1WES0
A0A026W9G8
+ More
A0A0C9RCY2 A0A067RAE9 E2ACW1 A0A151J3G5 A0A0J7KX56 A0A195F7A0 A0A195B925 H9J8S9 A0A195D087 E2BFN9 A0A2A3EMG5 A0A0N0U2M9 E9J2W8 A0A2P8ZDR8 A0A087ZN09 A0A151WYH3 K7J8M2 A0A0T6AV00 A0A232F0D6 A0A1Y1KZI8 D6WM60 A0A1W4WWK5 A0A069DV12 T1HTG8 A0A224XLQ9 A0A0P4VMH0 A0A1S3D4X9 A0A3Q0IY14 A0A158NWN3 A0A0L7R405 A0A2J7RGD5 A0A2J7RGC6 A0A1B6CH82 A0A146LVI8 A0A182PG13 A0A182RAY3 A0A182YDP3 A0A182VUI0 A0A182U7K3 A0A182X0S2 A0A1S4H0L1 Q7PVH1 A0A182M8I0 A0A182V9A8 A0A182IUV3 A0A182NU60 A0A336LWA8 A0A1Q3EXM7 A0A182QAZ9 A0A182L6F5 A0A1Q3EXU2 A0A084VWH1 W5J9Z1 A0A182F4R2 A0A182HGF9 A0A1S4FI55 A0A182K420 A0A182GAE9 A0A182G7I0 A0A1J1IGQ9 Q170B4 B0WRA4 A0A1I8QES0 A0A1I8MYS6 A0A0K8V6I6 B0X2C2 A0A1B0A1J7 A0A1B0G508 A0A1A9Y4D2 A0A1B0BG35 A0A0A1WER6 A0A0L0C8Y6 Q5NDL5 W8C3X0 A0A0M4E3K5 B4NWP0 Q5NDL4 B4GSK1 A0A0J9QUB8 B4Q838 B3N9K9 B4LUD1 E0VER0 B4KGV4 M9PBV8 Q9VQB7 A0A1W4UUC9 A0A0P4WRY1 A0A1Y1KX70 J9JUW5 B3MLI9 B4MTZ2
A0A0C9RCY2 A0A067RAE9 E2ACW1 A0A151J3G5 A0A0J7KX56 A0A195F7A0 A0A195B925 H9J8S9 A0A195D087 E2BFN9 A0A2A3EMG5 A0A0N0U2M9 E9J2W8 A0A2P8ZDR8 A0A087ZN09 A0A151WYH3 K7J8M2 A0A0T6AV00 A0A232F0D6 A0A1Y1KZI8 D6WM60 A0A1W4WWK5 A0A069DV12 T1HTG8 A0A224XLQ9 A0A0P4VMH0 A0A1S3D4X9 A0A3Q0IY14 A0A158NWN3 A0A0L7R405 A0A2J7RGD5 A0A2J7RGC6 A0A1B6CH82 A0A146LVI8 A0A182PG13 A0A182RAY3 A0A182YDP3 A0A182VUI0 A0A182U7K3 A0A182X0S2 A0A1S4H0L1 Q7PVH1 A0A182M8I0 A0A182V9A8 A0A182IUV3 A0A182NU60 A0A336LWA8 A0A1Q3EXM7 A0A182QAZ9 A0A182L6F5 A0A1Q3EXU2 A0A084VWH1 W5J9Z1 A0A182F4R2 A0A182HGF9 A0A1S4FI55 A0A182K420 A0A182GAE9 A0A182G7I0 A0A1J1IGQ9 Q170B4 B0WRA4 A0A1I8QES0 A0A1I8MYS6 A0A0K8V6I6 B0X2C2 A0A1B0A1J7 A0A1B0G508 A0A1A9Y4D2 A0A1B0BG35 A0A0A1WER6 A0A0L0C8Y6 Q5NDL5 W8C3X0 A0A0M4E3K5 B4NWP0 Q5NDL4 B4GSK1 A0A0J9QUB8 B4Q838 B3N9K9 B4LUD1 E0VER0 B4KGV4 M9PBV8 Q9VQB7 A0A1W4UUC9 A0A0P4WRY1 A0A1Y1KX70 J9JUW5 B3MLI9 B4MTZ2
EC Number
2.4.1.255
Pubmed
26354079
28756777
24508170
30249741
24845553
20798317
+ More
19121390 21282665 29403074 20075255 28648823 28004739 18362917 19820115 26334808 27129103 21347285 26823975 25244985 12364791 20966253 24438588 20920257 23761445 26483478 17510324 25315136 25830018 26108605 15632085 17994087 23185243 24495485 17550304 22936249 18057021 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22158438 12537569
19121390 21282665 29403074 20075255 28648823 28004739 18362917 19820115 26334808 27129103 21347285 26823975 25244985 12364791 20966253 24438588 20920257 23761445 26483478 17510324 25315136 25830018 26108605 15632085 17994087 23185243 24495485 17550304 22936249 18057021 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22158438 12537569
EMBL
NWSH01000672
PCG74896.1
KQ460398
KPJ15151.1
KQ459601
KPI93835.1
+ More
KZ149895 PZC78789.1 ODYU01008149 SOQ51517.1 KK107347 QOIP01000007 EZA52321.1 RLU20673.1 GBYB01014444 JAG84211.1 KK852595 KDR20615.1 GL438586 EFN68718.1 KQ980278 KYN16979.1 LBMM01002229 KMQ95102.1 KQ981744 KYN36318.1 KQ976558 KYM80732.1 BABH01005101 BABH01005102 BABH01005103 KQ977012 KYN06333.1 GL448037 EFN85561.1 KZ288211 PBC32897.1 KQ436245 KOX67308.1 GL768054 EFZ12831.1 PYGN01000087 PSN54636.1 KQ982650 KYQ52929.1 AAZX01000971 AAZX01016287 LJIG01022744 KRT78940.1 NNAY01001362 OXU24236.1 GEZM01073428 JAV65105.1 KQ971343 EFA03354.1 GBGD01001228 JAC87661.1 ACPB03016020 GFTR01006894 JAW09532.1 GDKW01002597 JAI53998.1 ADTU01028242 KQ414661 KOC65578.1 NEVH01004401 PNF39883.1 PNF39882.1 GEDC01028744 GEDC01024760 GEDC01024546 GEDC01004390 GEDC01003895 JAS08554.1 JAS12538.1 JAS12752.1 JAS32908.1 JAS33403.1 GDHC01007933 JAQ10696.1 AAAB01008984 EAA14911.4 AXCM01002460 UFQS01000240 UFQT01000240 SSX01877.1 SSX22254.1 GFDL01014981 JAV20064.1 AXCN02000773 GFDL01015017 JAV20028.1 ATLV01017586 KE525174 KFB42315.1 ADMH02001962 ETN60228.1 APCN01004224 JXUM01050896 KQ561666 KXJ77865.1 JXUM01047550 KQ561524 KXJ78286.1 CVRI01000048 CRK98954.1 CH477477 EAT40278.1 DS232053 EDS33288.1 GDHF01017772 JAI34542.1 DS232286 EDS39145.1 CCAG010023642 JXJN01013737 GBXI01017111 JAC97180.1 JRES01000753 KNC28707.1 AJ868231 CH379061 CAI30566.1 EAL33029.1 GAMC01002597 JAC03959.1 CP012523 ALC38221.1 CM000157 EDW87382.1 AJ868232 CAI30567.1 CH479189 EDW25360.1 CM002910 KMY87667.1 CM000361 EDX03460.1 CH954177 EDV57466.1 CH940649 EDW64117.1 KRF81435.1 DS235093 EEB11866.1 CH933807 EDW11154.1 AE014134 AGB92471.1 AB675601 AY058292 GDRN01038051 GDRN01038050 JAI67269.1 GEZM01073429 JAV65101.1 ABLF02037452 CH902620 EDV31738.1 CH963852 EDW75581.1
KZ149895 PZC78789.1 ODYU01008149 SOQ51517.1 KK107347 QOIP01000007 EZA52321.1 RLU20673.1 GBYB01014444 JAG84211.1 KK852595 KDR20615.1 GL438586 EFN68718.1 KQ980278 KYN16979.1 LBMM01002229 KMQ95102.1 KQ981744 KYN36318.1 KQ976558 KYM80732.1 BABH01005101 BABH01005102 BABH01005103 KQ977012 KYN06333.1 GL448037 EFN85561.1 KZ288211 PBC32897.1 KQ436245 KOX67308.1 GL768054 EFZ12831.1 PYGN01000087 PSN54636.1 KQ982650 KYQ52929.1 AAZX01000971 AAZX01016287 LJIG01022744 KRT78940.1 NNAY01001362 OXU24236.1 GEZM01073428 JAV65105.1 KQ971343 EFA03354.1 GBGD01001228 JAC87661.1 ACPB03016020 GFTR01006894 JAW09532.1 GDKW01002597 JAI53998.1 ADTU01028242 KQ414661 KOC65578.1 NEVH01004401 PNF39883.1 PNF39882.1 GEDC01028744 GEDC01024760 GEDC01024546 GEDC01004390 GEDC01003895 JAS08554.1 JAS12538.1 JAS12752.1 JAS32908.1 JAS33403.1 GDHC01007933 JAQ10696.1 AAAB01008984 EAA14911.4 AXCM01002460 UFQS01000240 UFQT01000240 SSX01877.1 SSX22254.1 GFDL01014981 JAV20064.1 AXCN02000773 GFDL01015017 JAV20028.1 ATLV01017586 KE525174 KFB42315.1 ADMH02001962 ETN60228.1 APCN01004224 JXUM01050896 KQ561666 KXJ77865.1 JXUM01047550 KQ561524 KXJ78286.1 CVRI01000048 CRK98954.1 CH477477 EAT40278.1 DS232053 EDS33288.1 GDHF01017772 JAI34542.1 DS232286 EDS39145.1 CCAG010023642 JXJN01013737 GBXI01017111 JAC97180.1 JRES01000753 KNC28707.1 AJ868231 CH379061 CAI30566.1 EAL33029.1 GAMC01002597 JAC03959.1 CP012523 ALC38221.1 CM000157 EDW87382.1 AJ868232 CAI30567.1 CH479189 EDW25360.1 CM002910 KMY87667.1 CM000361 EDX03460.1 CH954177 EDV57466.1 CH940649 EDW64117.1 KRF81435.1 DS235093 EEB11866.1 CH933807 EDW11154.1 AE014134 AGB92471.1 AB675601 AY058292 GDRN01038051 GDRN01038050 JAI67269.1 GEZM01073429 JAV65101.1 ABLF02037452 CH902620 EDV31738.1 CH963852 EDW75581.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000053097
UP000279307
UP000027135
+ More
UP000000311 UP000078492 UP000036403 UP000078541 UP000078540 UP000005204 UP000078542 UP000008237 UP000242457 UP000053105 UP000245037 UP000005203 UP000075809 UP000002358 UP000215335 UP000007266 UP000192223 UP000015103 UP000079169 UP000005205 UP000053825 UP000235965 UP000075885 UP000075900 UP000076408 UP000075920 UP000075902 UP000076407 UP000007062 UP000075883 UP000075903 UP000075880 UP000075884 UP000075886 UP000075882 UP000030765 UP000000673 UP000069272 UP000075840 UP000075881 UP000069940 UP000249989 UP000183832 UP000008820 UP000002320 UP000095300 UP000095301 UP000092445 UP000092444 UP000092443 UP000092460 UP000037069 UP000001819 UP000092553 UP000002282 UP000008744 UP000000304 UP000008711 UP000008792 UP000009046 UP000009192 UP000000803 UP000192221 UP000007819 UP000007801 UP000007798
UP000000311 UP000078492 UP000036403 UP000078541 UP000078540 UP000005204 UP000078542 UP000008237 UP000242457 UP000053105 UP000245037 UP000005203 UP000075809 UP000002358 UP000215335 UP000007266 UP000192223 UP000015103 UP000079169 UP000005205 UP000053825 UP000235965 UP000075885 UP000075900 UP000076408 UP000075920 UP000075902 UP000076407 UP000007062 UP000075883 UP000075903 UP000075880 UP000075884 UP000075886 UP000075882 UP000030765 UP000000673 UP000069272 UP000075840 UP000075881 UP000069940 UP000249989 UP000183832 UP000008820 UP000002320 UP000095300 UP000095301 UP000092445 UP000092444 UP000092443 UP000092460 UP000037069 UP000001819 UP000092553 UP000002282 UP000008744 UP000000304 UP000008711 UP000008792 UP000009046 UP000009192 UP000000803 UP000192221 UP000007819 UP000007801 UP000007798
ProteinModelPortal
A0A2A4JSY7
A0A194RGU5
A0A194PRS1
A0A2W1BUZ4
A0A2H1WES0
A0A026W9G8
+ More
A0A0C9RCY2 A0A067RAE9 E2ACW1 A0A151J3G5 A0A0J7KX56 A0A195F7A0 A0A195B925 H9J8S9 A0A195D087 E2BFN9 A0A2A3EMG5 A0A0N0U2M9 E9J2W8 A0A2P8ZDR8 A0A087ZN09 A0A151WYH3 K7J8M2 A0A0T6AV00 A0A232F0D6 A0A1Y1KZI8 D6WM60 A0A1W4WWK5 A0A069DV12 T1HTG8 A0A224XLQ9 A0A0P4VMH0 A0A1S3D4X9 A0A3Q0IY14 A0A158NWN3 A0A0L7R405 A0A2J7RGD5 A0A2J7RGC6 A0A1B6CH82 A0A146LVI8 A0A182PG13 A0A182RAY3 A0A182YDP3 A0A182VUI0 A0A182U7K3 A0A182X0S2 A0A1S4H0L1 Q7PVH1 A0A182M8I0 A0A182V9A8 A0A182IUV3 A0A182NU60 A0A336LWA8 A0A1Q3EXM7 A0A182QAZ9 A0A182L6F5 A0A1Q3EXU2 A0A084VWH1 W5J9Z1 A0A182F4R2 A0A182HGF9 A0A1S4FI55 A0A182K420 A0A182GAE9 A0A182G7I0 A0A1J1IGQ9 Q170B4 B0WRA4 A0A1I8QES0 A0A1I8MYS6 A0A0K8V6I6 B0X2C2 A0A1B0A1J7 A0A1B0G508 A0A1A9Y4D2 A0A1B0BG35 A0A0A1WER6 A0A0L0C8Y6 Q5NDL5 W8C3X0 A0A0M4E3K5 B4NWP0 Q5NDL4 B4GSK1 A0A0J9QUB8 B4Q838 B3N9K9 B4LUD1 E0VER0 B4KGV4 M9PBV8 Q9VQB7 A0A1W4UUC9 A0A0P4WRY1 A0A1Y1KX70 J9JUW5 B3MLI9 B4MTZ2
A0A0C9RCY2 A0A067RAE9 E2ACW1 A0A151J3G5 A0A0J7KX56 A0A195F7A0 A0A195B925 H9J8S9 A0A195D087 E2BFN9 A0A2A3EMG5 A0A0N0U2M9 E9J2W8 A0A2P8ZDR8 A0A087ZN09 A0A151WYH3 K7J8M2 A0A0T6AV00 A0A232F0D6 A0A1Y1KZI8 D6WM60 A0A1W4WWK5 A0A069DV12 T1HTG8 A0A224XLQ9 A0A0P4VMH0 A0A1S3D4X9 A0A3Q0IY14 A0A158NWN3 A0A0L7R405 A0A2J7RGD5 A0A2J7RGC6 A0A1B6CH82 A0A146LVI8 A0A182PG13 A0A182RAY3 A0A182YDP3 A0A182VUI0 A0A182U7K3 A0A182X0S2 A0A1S4H0L1 Q7PVH1 A0A182M8I0 A0A182V9A8 A0A182IUV3 A0A182NU60 A0A336LWA8 A0A1Q3EXM7 A0A182QAZ9 A0A182L6F5 A0A1Q3EXU2 A0A084VWH1 W5J9Z1 A0A182F4R2 A0A182HGF9 A0A1S4FI55 A0A182K420 A0A182GAE9 A0A182G7I0 A0A1J1IGQ9 Q170B4 B0WRA4 A0A1I8QES0 A0A1I8MYS6 A0A0K8V6I6 B0X2C2 A0A1B0A1J7 A0A1B0G508 A0A1A9Y4D2 A0A1B0BG35 A0A0A1WER6 A0A0L0C8Y6 Q5NDL5 W8C3X0 A0A0M4E3K5 B4NWP0 Q5NDL4 B4GSK1 A0A0J9QUB8 B4Q838 B3N9K9 B4LUD1 E0VER0 B4KGV4 M9PBV8 Q9VQB7 A0A1W4UUC9 A0A0P4WRY1 A0A1Y1KX70 J9JUW5 B3MLI9 B4MTZ2
Ontologies
GO
PANTHER
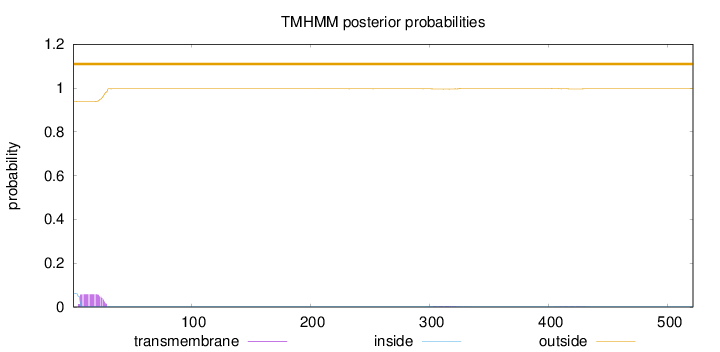
Topology
Subcellular location
Endoplasmic reticulum lumen
SignalP
Position: 1 - 19,
Likelihood: 0.966511
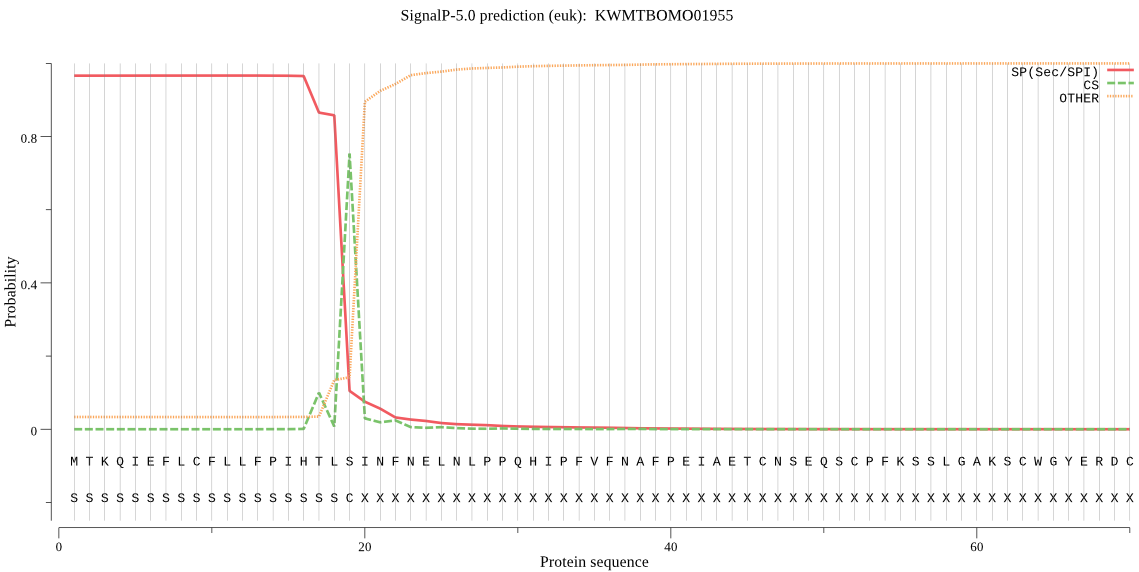
Length:
521
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.29043
Exp number, first 60 AAs:
1.1674
Total prob of N-in:
0.06088
outside
1 - 521
Population Genetic Test Statistics
Pi
195.613799
Theta
180.543879
Tajima's D
0.829446
CLR
0.35875
CSRT
0.605169741512924
Interpretation
Uncertain
