Gene
KWMTBOMO01945
Pre Gene Modal
BGIBMGA006224
Annotation
PREDICTED:_transmembrane_protein_189_isoform_X1_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 2.578
Sequence
CDS
ATGGCTTTGCCGTTACCCATATCGCCTTGTGCTAAAAGAGGGGACGGTATCAGTGCTTTGGGCTGCGAGCGATTCGAATACGCAAGCACTATGGCCACAACCCTGTTAGCGCCGGTTAAAACTGAACGTGAGATCCTAGAGCATTCGATGTCGGAAGATGATCCTAACGCAAATACACAATTATCAAGTCAAGTTGGAACCCAACCACGTTGGGGTCCAAGGCATAAAGGCGCACAAGAACTTGCTGAATTGTATAGTCCAGGCAAGCGATTGCAGGAATGGGTATGTGTGGTATTATGTTGCACACTATGTGCCTCTGCCGGAATTTTAATGATCAGATATGTTCGGGCTGATGCTTCACTGGTACTGGCAGCTCTTGCAGGGGTCTTAACAGCAGATTTTGCTTCTGGAGTTGTGCACTGGGCAGCTGACACTTGGGGAGCAGTTGATCTACCAATAATAGGCAAAAACTTTCTCCGACCATTTCGAGAGCACCACATCGATCCTACATCAATCACCCGTCATGACTTTATTGAGACCAATGGTGATAATTTTGCTATAACGATACCTGTACTTGCACGAATAGTATGGCAACTAGTTACGTACGAAGAAGGAAAAGTACATGACCAATTTCATTGGATTGCATATTGGTATTTGTGCTGTATATTCGTGGCTATGACGAATCAGATTCACAAATGGTCGCACACATATTTTGGGTTGCCATCATGGGTGGTGTGGTTACAAGAGTGGCACATTGTGCTGCCTCGTCGCCATCATCGTATACATCACGTCGCACCGCACGAGACTTACTTTTGTATAACCACTGGCTGGCTTAACTGGCCATTAGAAAAACTTCACTTTTGGTCGACTTTAGAGACAATAATAGAAGCTGTAACTGGATGTAGACCACGAGCAGACGACCTTAAATGGGCACAAAAACGTTCCTAG
Protein
MALPLPISPCAKRGDGISALGCERFEYASTMATTLLAPVKTEREILEHSMSEDDPNANTQLSSQVGTQPRWGPRHKGAQELAELYSPGKRLQEWVCVVLCCTLCASAGILMIRYVRADASLVLAALAGVLTADFASGVVHWAADTWGAVDLPIIGKNFLRPFREHHIDPTSITRHDFIETNGDNFAITIPVLARIVWQLVTYEEGKVHDQFHWIAYWYLCCIFVAMTNQIHKWSHTYFGLPSWVVWLQEWHIVLPRRHHRIHHVAPHETYFCITTGWLNWPLEKLHFWSTLETIIEAVTGCRPRADDLKWAQKRS
Summary
Uniprot
H9J9N1
A0A194RBF5
A0A194PRT1
A0A212ETS7
A0A2A4K4E1
A0A2A4K3Y8
+ More
A0A1Y1K217 A0A0L7LKH0 A0A0M8ZWY6 D6WJA6 A0A0J7NU39 E2AH55 E2C362 A0A2J7Q1N1 A0A026VVG2 A0A195FSP9 A0A2A3E7K3 A0A087ZV95 A0A195BKH0 A0A158NA45 A0A195DLN9 A0A195CWD2 A0A067QNV2 E9IXM8 F4X1E9 A0A151WWN7 A0A2P8XU89 A0A232EYF6 N6UB96 A0A182WGF5 A0A182Q206 A0A1B6D822 A0A182IQS6 A0A182MGV5 A0A1L8DSG0 A0A182F109 A0A182VDV7 A0A2W1C1G0 A0A182Y0K5 A0A1B6L8T3 A0A182LMA1 A0A336LYN7 A0A182NBN0 A0A182K6I3 W5J8I9 A0A1B6JK45 A0A0P4WH69 A0A0L7RG65 Q7Q1N6 A0A1I8NX02 A0A1I8MLN3 A0A034VBB6 A0A0K8WKU9 A0A0K8U0E0 A0A0L0C4V2 A0A0A1X1N5 K7IQQ7 A0A0A1WP99 A0A1J1ISL8 Q17LY2 A0A1S4EY35 A0A224ZBF3 A0A1A9UEY2 L7ME04 A0A1A9YF93 A0A1B0AH72 A0A1B0BX05 V5H9R1 A0A084WU94 A0A1A9WHG1 A0A2R5LK94 A0A293M839 W8BD85 W8C0T1 A0A182XFS8 A0A182HI63 A0A2P2I5P8 B4KEW2 A0A210PT33 A0A0R3NUI4 A0A0P8XFI4 B4GT61 B3MKV7 A0A3B0KCN5 Q29KL3 B4JQ98 A0A023F5Y2 A0A1W4VXW3 T1HYC9 B4M9M6 B4PBI3 B4I639 B4QAH5 K1P739 Q9V3B5 B3NKZ8 B4N7R2 A0A146LSV0 A0A0K8SDX3 A0A0A9W1H0
A0A1Y1K217 A0A0L7LKH0 A0A0M8ZWY6 D6WJA6 A0A0J7NU39 E2AH55 E2C362 A0A2J7Q1N1 A0A026VVG2 A0A195FSP9 A0A2A3E7K3 A0A087ZV95 A0A195BKH0 A0A158NA45 A0A195DLN9 A0A195CWD2 A0A067QNV2 E9IXM8 F4X1E9 A0A151WWN7 A0A2P8XU89 A0A232EYF6 N6UB96 A0A182WGF5 A0A182Q206 A0A1B6D822 A0A182IQS6 A0A182MGV5 A0A1L8DSG0 A0A182F109 A0A182VDV7 A0A2W1C1G0 A0A182Y0K5 A0A1B6L8T3 A0A182LMA1 A0A336LYN7 A0A182NBN0 A0A182K6I3 W5J8I9 A0A1B6JK45 A0A0P4WH69 A0A0L7RG65 Q7Q1N6 A0A1I8NX02 A0A1I8MLN3 A0A034VBB6 A0A0K8WKU9 A0A0K8U0E0 A0A0L0C4V2 A0A0A1X1N5 K7IQQ7 A0A0A1WP99 A0A1J1ISL8 Q17LY2 A0A1S4EY35 A0A224ZBF3 A0A1A9UEY2 L7ME04 A0A1A9YF93 A0A1B0AH72 A0A1B0BX05 V5H9R1 A0A084WU94 A0A1A9WHG1 A0A2R5LK94 A0A293M839 W8BD85 W8C0T1 A0A182XFS8 A0A182HI63 A0A2P2I5P8 B4KEW2 A0A210PT33 A0A0R3NUI4 A0A0P8XFI4 B4GT61 B3MKV7 A0A3B0KCN5 Q29KL3 B4JQ98 A0A023F5Y2 A0A1W4VXW3 T1HYC9 B4M9M6 B4PBI3 B4I639 B4QAH5 K1P739 Q9V3B5 B3NKZ8 B4N7R2 A0A146LSV0 A0A0K8SDX3 A0A0A9W1H0
Pubmed
19121390
26354079
22118469
28004739
26227816
18362917
+ More
19820115 20798317 24508170 30249741 21347285 24845553 21282665 21719571 29403074 28648823 23537049 28756777 25244985 20966253 20920257 23761445 12364791 25315136 25348373 26108605 25830018 20075255 17510324 28797301 25576852 25765539 24438588 24495485 17994087 18057021 28812685 15632085 23185243 25474469 17550304 22936249 22992520 11076860 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 25401762
19820115 20798317 24508170 30249741 21347285 24845553 21282665 21719571 29403074 28648823 23537049 28756777 25244985 20966253 20920257 23761445 12364791 25315136 25348373 26108605 25830018 20075255 17510324 28797301 25576852 25765539 24438588 24495485 17994087 18057021 28812685 15632085 23185243 25474469 17550304 22936249 22992520 11076860 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26823975 25401762
EMBL
BABH01005081
BABH01005082
KQ460398
KPJ15163.1
KQ459601
KPI93845.1
+ More
AGBW02012530 OWR44905.1 NWSH01000172 PCG78778.1 PCG78779.1 GEZM01099226 JAV53426.1 JTDY01000738 KOB76053.1 KQ435830 KOX71746.1 KQ971343 EFA03149.1 LBMM01001664 KMQ95925.1 GL439483 EFN67172.1 GL452292 EFN77594.1 NEVH01019394 PNF22491.1 KK107796 QOIP01000013 EZA47625.1 RLU15229.1 KQ981305 KYN42929.1 KZ288348 PBC27484.1 KQ976455 KYM85110.1 ADTU01009965 KQ980734 KYN13793.1 KQ977276 KYN04464.1 KK853129 KDR10990.1 GL766762 EFZ14650.1 GL888529 EGI59744.1 KQ982686 KYQ52324.1 PYGN01001356 PSN35464.1 NNAY01001691 OXU23208.1 APGK01041724 KB740996 ENN75912.1 AXCN02000679 GEDC01015464 GEDC01011651 GEDC01011073 GEDC01006853 JAS21834.1 JAS25647.1 JAS26225.1 JAS30445.1 AXCM01003195 GFDF01004705 JAV09379.1 KZ149895 PZC78806.1 GEBQ01019860 JAT20117.1 UFQS01000320 UFQT01000320 SSX02828.1 SSX23196.1 ADMH02001955 ETN60281.1 GECU01008118 JAS99588.1 GDRN01052498 JAI66292.1 KQ414598 KOC69823.1 AAAB01008980 EAA14566.3 GAKP01018362 GAKP01018360 GAKP01018356 JAC40596.1 GDHF01013454 GDHF01000541 JAI38860.1 JAI51773.1 GDHF01032554 JAI19760.1 JRES01000914 KNC27277.1 GBXI01009622 JAD04670.1 GBXI01014039 JAD00253.1 CVRI01000055 CRL01521.1 CH477210 EAT47689.1 GFPF01013084 MAA24230.1 GACK01002824 JAA62210.1 JXJN01022022 GANP01010554 JAB73914.1 ATLV01027020 KE525421 KFB53788.1 GGLE01005773 MBY09899.1 GFWV01010571 MAA35300.1 GAMC01007296 GAMC01007295 JAB99260.1 GAMC01007294 JAB99261.1 APCN01005628 IACF01003753 LAB69363.1 CH933807 EDW11931.1 KRG02975.1 KRG02976.1 KRG02977.1 NEDP02005518 OWF39657.1 CH379061 KRT04786.1 KRT04787.1 CH902620 KPU73519.1 CH479189 EDW25570.1 EDV31638.1 KPU73518.1 KPU73520.1 KPU73521.1 OUUW01000010 SPP85980.1 EAL33161.2 CH916372 EDV99078.1 GBBI01002109 JAC16603.1 ACPB03007951 CH940654 EDW57902.1 KRF78003.1 KRF78004.1 KRF78005.1 KRF78006.1 CM000158 EDW90497.1 KRJ99218.1 KRJ99219.1 CH480822 EDW55845.1 CM000361 CM002910 EDX05592.1 KMY91134.1 JH816441 EKC19452.1 AF152361 AE014134 BT029029 BT126266 AAF00151.1 AAF53878.1 ABJ16962.1 ADV37105.1 AEB39627.1 AGB93155.1 AGB93156.1 CH954179 EDV54576.1 KQS61844.1 CH964214 EDW80401.1 KRF99275.1 GDHC01008787 JAQ09842.1 GBRD01014517 JAG51309.1 GBHO01041960 GBHO01041956 JAG01644.1 JAG01648.1
AGBW02012530 OWR44905.1 NWSH01000172 PCG78778.1 PCG78779.1 GEZM01099226 JAV53426.1 JTDY01000738 KOB76053.1 KQ435830 KOX71746.1 KQ971343 EFA03149.1 LBMM01001664 KMQ95925.1 GL439483 EFN67172.1 GL452292 EFN77594.1 NEVH01019394 PNF22491.1 KK107796 QOIP01000013 EZA47625.1 RLU15229.1 KQ981305 KYN42929.1 KZ288348 PBC27484.1 KQ976455 KYM85110.1 ADTU01009965 KQ980734 KYN13793.1 KQ977276 KYN04464.1 KK853129 KDR10990.1 GL766762 EFZ14650.1 GL888529 EGI59744.1 KQ982686 KYQ52324.1 PYGN01001356 PSN35464.1 NNAY01001691 OXU23208.1 APGK01041724 KB740996 ENN75912.1 AXCN02000679 GEDC01015464 GEDC01011651 GEDC01011073 GEDC01006853 JAS21834.1 JAS25647.1 JAS26225.1 JAS30445.1 AXCM01003195 GFDF01004705 JAV09379.1 KZ149895 PZC78806.1 GEBQ01019860 JAT20117.1 UFQS01000320 UFQT01000320 SSX02828.1 SSX23196.1 ADMH02001955 ETN60281.1 GECU01008118 JAS99588.1 GDRN01052498 JAI66292.1 KQ414598 KOC69823.1 AAAB01008980 EAA14566.3 GAKP01018362 GAKP01018360 GAKP01018356 JAC40596.1 GDHF01013454 GDHF01000541 JAI38860.1 JAI51773.1 GDHF01032554 JAI19760.1 JRES01000914 KNC27277.1 GBXI01009622 JAD04670.1 GBXI01014039 JAD00253.1 CVRI01000055 CRL01521.1 CH477210 EAT47689.1 GFPF01013084 MAA24230.1 GACK01002824 JAA62210.1 JXJN01022022 GANP01010554 JAB73914.1 ATLV01027020 KE525421 KFB53788.1 GGLE01005773 MBY09899.1 GFWV01010571 MAA35300.1 GAMC01007296 GAMC01007295 JAB99260.1 GAMC01007294 JAB99261.1 APCN01005628 IACF01003753 LAB69363.1 CH933807 EDW11931.1 KRG02975.1 KRG02976.1 KRG02977.1 NEDP02005518 OWF39657.1 CH379061 KRT04786.1 KRT04787.1 CH902620 KPU73519.1 CH479189 EDW25570.1 EDV31638.1 KPU73518.1 KPU73520.1 KPU73521.1 OUUW01000010 SPP85980.1 EAL33161.2 CH916372 EDV99078.1 GBBI01002109 JAC16603.1 ACPB03007951 CH940654 EDW57902.1 KRF78003.1 KRF78004.1 KRF78005.1 KRF78006.1 CM000158 EDW90497.1 KRJ99218.1 KRJ99219.1 CH480822 EDW55845.1 CM000361 CM002910 EDX05592.1 KMY91134.1 JH816441 EKC19452.1 AF152361 AE014134 BT029029 BT126266 AAF00151.1 AAF53878.1 ABJ16962.1 ADV37105.1 AEB39627.1 AGB93155.1 AGB93156.1 CH954179 EDV54576.1 KQS61844.1 CH964214 EDW80401.1 KRF99275.1 GDHC01008787 JAQ09842.1 GBRD01014517 JAG51309.1 GBHO01041960 GBHO01041956 JAG01644.1 JAG01648.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000218220
UP000037510
+ More
UP000053105 UP000007266 UP000036403 UP000000311 UP000008237 UP000235965 UP000053097 UP000279307 UP000078541 UP000242457 UP000005203 UP000078540 UP000005205 UP000078492 UP000078542 UP000027135 UP000007755 UP000075809 UP000245037 UP000215335 UP000019118 UP000075920 UP000075886 UP000075880 UP000075883 UP000069272 UP000075903 UP000076408 UP000075882 UP000075884 UP000075881 UP000000673 UP000053825 UP000007062 UP000095300 UP000095301 UP000037069 UP000002358 UP000183832 UP000008820 UP000078200 UP000092443 UP000092445 UP000092460 UP000030765 UP000091820 UP000076407 UP000075840 UP000009192 UP000242188 UP000001819 UP000007801 UP000008744 UP000268350 UP000001070 UP000192221 UP000015103 UP000008792 UP000002282 UP000001292 UP000000304 UP000005408 UP000000803 UP000008711 UP000007798
UP000053105 UP000007266 UP000036403 UP000000311 UP000008237 UP000235965 UP000053097 UP000279307 UP000078541 UP000242457 UP000005203 UP000078540 UP000005205 UP000078492 UP000078542 UP000027135 UP000007755 UP000075809 UP000245037 UP000215335 UP000019118 UP000075920 UP000075886 UP000075880 UP000075883 UP000069272 UP000075903 UP000076408 UP000075882 UP000075884 UP000075881 UP000000673 UP000053825 UP000007062 UP000095300 UP000095301 UP000037069 UP000002358 UP000183832 UP000008820 UP000078200 UP000092443 UP000092445 UP000092460 UP000030765 UP000091820 UP000076407 UP000075840 UP000009192 UP000242188 UP000001819 UP000007801 UP000008744 UP000268350 UP000001070 UP000192221 UP000015103 UP000008792 UP000002282 UP000001292 UP000000304 UP000005408 UP000000803 UP000008711 UP000007798
Interpro
SUPFAM
SSF46561
SSF46561
ProteinModelPortal
H9J9N1
A0A194RBF5
A0A194PRT1
A0A212ETS7
A0A2A4K4E1
A0A2A4K3Y8
+ More
A0A1Y1K217 A0A0L7LKH0 A0A0M8ZWY6 D6WJA6 A0A0J7NU39 E2AH55 E2C362 A0A2J7Q1N1 A0A026VVG2 A0A195FSP9 A0A2A3E7K3 A0A087ZV95 A0A195BKH0 A0A158NA45 A0A195DLN9 A0A195CWD2 A0A067QNV2 E9IXM8 F4X1E9 A0A151WWN7 A0A2P8XU89 A0A232EYF6 N6UB96 A0A182WGF5 A0A182Q206 A0A1B6D822 A0A182IQS6 A0A182MGV5 A0A1L8DSG0 A0A182F109 A0A182VDV7 A0A2W1C1G0 A0A182Y0K5 A0A1B6L8T3 A0A182LMA1 A0A336LYN7 A0A182NBN0 A0A182K6I3 W5J8I9 A0A1B6JK45 A0A0P4WH69 A0A0L7RG65 Q7Q1N6 A0A1I8NX02 A0A1I8MLN3 A0A034VBB6 A0A0K8WKU9 A0A0K8U0E0 A0A0L0C4V2 A0A0A1X1N5 K7IQQ7 A0A0A1WP99 A0A1J1ISL8 Q17LY2 A0A1S4EY35 A0A224ZBF3 A0A1A9UEY2 L7ME04 A0A1A9YF93 A0A1B0AH72 A0A1B0BX05 V5H9R1 A0A084WU94 A0A1A9WHG1 A0A2R5LK94 A0A293M839 W8BD85 W8C0T1 A0A182XFS8 A0A182HI63 A0A2P2I5P8 B4KEW2 A0A210PT33 A0A0R3NUI4 A0A0P8XFI4 B4GT61 B3MKV7 A0A3B0KCN5 Q29KL3 B4JQ98 A0A023F5Y2 A0A1W4VXW3 T1HYC9 B4M9M6 B4PBI3 B4I639 B4QAH5 K1P739 Q9V3B5 B3NKZ8 B4N7R2 A0A146LSV0 A0A0K8SDX3 A0A0A9W1H0
A0A1Y1K217 A0A0L7LKH0 A0A0M8ZWY6 D6WJA6 A0A0J7NU39 E2AH55 E2C362 A0A2J7Q1N1 A0A026VVG2 A0A195FSP9 A0A2A3E7K3 A0A087ZV95 A0A195BKH0 A0A158NA45 A0A195DLN9 A0A195CWD2 A0A067QNV2 E9IXM8 F4X1E9 A0A151WWN7 A0A2P8XU89 A0A232EYF6 N6UB96 A0A182WGF5 A0A182Q206 A0A1B6D822 A0A182IQS6 A0A182MGV5 A0A1L8DSG0 A0A182F109 A0A182VDV7 A0A2W1C1G0 A0A182Y0K5 A0A1B6L8T3 A0A182LMA1 A0A336LYN7 A0A182NBN0 A0A182K6I3 W5J8I9 A0A1B6JK45 A0A0P4WH69 A0A0L7RG65 Q7Q1N6 A0A1I8NX02 A0A1I8MLN3 A0A034VBB6 A0A0K8WKU9 A0A0K8U0E0 A0A0L0C4V2 A0A0A1X1N5 K7IQQ7 A0A0A1WP99 A0A1J1ISL8 Q17LY2 A0A1S4EY35 A0A224ZBF3 A0A1A9UEY2 L7ME04 A0A1A9YF93 A0A1B0AH72 A0A1B0BX05 V5H9R1 A0A084WU94 A0A1A9WHG1 A0A2R5LK94 A0A293M839 W8BD85 W8C0T1 A0A182XFS8 A0A182HI63 A0A2P2I5P8 B4KEW2 A0A210PT33 A0A0R3NUI4 A0A0P8XFI4 B4GT61 B3MKV7 A0A3B0KCN5 Q29KL3 B4JQ98 A0A023F5Y2 A0A1W4VXW3 T1HYC9 B4M9M6 B4PBI3 B4I639 B4QAH5 K1P739 Q9V3B5 B3NKZ8 B4N7R2 A0A146LSV0 A0A0K8SDX3 A0A0A9W1H0
Ontologies
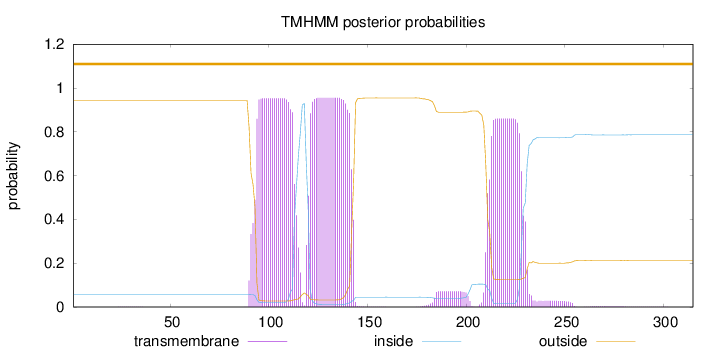
Topology
Length:
315
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
60.10436
Exp number, first 60 AAs:
0.0024
Total prob of N-in:
0.05690
outside
1 - 315
Population Genetic Test Statistics
Pi
239.883733
Theta
180.929119
Tajima's D
1.122914
CLR
0.92663
CSRT
0.690465476726164
Interpretation
Uncertain
