Gene
KWMTBOMO01944
Pre Gene Modal
BGIBMGA005927
Annotation
PREDICTED:_N-terminal_Xaa-Pro-Lys_N-methyltransferase_1-A-like_[Papilio_machaon]
Full name
Alpha N-terminal protein methyltransferase 1
Alternative Name
X-Pro-Lys N-terminal protein methyltransferase 1
Location in the cell
Cytoplasmic Reliability : 1.994
Sequence
CDS
ATGAGTAATTTAGATTTTGATGATAACGACATAAGTTATGAAAGTGCCCAAAAATATTGGTCAGATATTCCTGCAACTGTTGATGGAGTTCTCGGTGGCTTTGGATTTATATCTGACATAGACATAGAAGGATCAAGAATTTTCCTTGAAAGTCTTTTATCGTGTAATAAAGCTCCATGCACTGATTTAGCACTTGACTGTGGTGCTGGTATAGGAAGGATAACAAAGAACTTACTATTGCCATACTTTCAATTAGTGGACGTTGTGGAGCCCGATAGTAAATTCATTAAAACAATAAAAAATTATGTTGGTGAACACCAATCAAAGATTGACACACTATACGAAGTTGGTTTACAAGATTTCAATCCTGTCAAAAGATATGATGTTATTTGGAACCAGTGGGTTCTAGGATATCTACTGGATCAGGATTTAATAGCTTACTTAAAACGCTGCAGAAATGCTTTAAAGACCAATGGAGTCATTGTAGTAAAAGAAAACGTAACTTCATCAGGTAAAAGTGAAAAGGATGAAACAGATTCCTCAGTTACACGATCACTACAACAGTTTCTTAGTATTTTTAAACAATCAAAACTGAAGAGATTAAAACAGTGTAAGCAAACTAATTTCCCTAATGGAATTTACCCTGTTTATATGTTTGCATTAGTTCCTATACAGGAAGATTATAATTCTGAGGATTTAAAGGTATAA
Protein
MSNLDFDDNDISYESAQKYWSDIPATVDGVLGGFGFISDIDIEGSRIFLESLLSCNKAPCTDLALDCGAGIGRITKNLLLPYFQLVDVVEPDSKFIKTIKNYVGEHQSKIDTLYEVGLQDFNPVKRYDVIWNQWVLGYLLDQDLIAYLKRCRNALKTNGVIVVKENVTSSGKSEKDETDSSVTRSLQQFLSIFKQSKLKRLKQCKQTNFPNGIYPVYMFALVPIQEDYNSEDLKV
Summary
Description
Alpha-N-methyltransferase that methylates the N-terminus of target proteins containing the N-terminal motif [Ala/Pro/Ser]-Pro-Lys when the initiator Met is cleaved. Specifically catalyzes mono-, di- or tri-methylation of exposed alpha-amino group of Ala or Ser residue in the [Ala/Ser]-Pro-Lys motif and mono- or di-methylation of Pro in the Pro-Pro-Lys motif (By similarity).
Catalytic Activity
N-terminal L-alanyl-L-prolyl-L-lysyl-[protein] + 3 S-adenosyl-L-methionine = 3 H(+) + N-terminal N,N,N-trimethyl-L-alanyl-L-prolyl-L-lysyl-[protein] + 3 S-adenosyl-L-homocysteine
N-terminal L-seryl-L-prolyl-L-lysyl-[protein] + 3 S-adenosyl-L-methionine = 3 H(+) + N-terminal N,N,N-trimethyl-L-seryl-L-prolyl-L-lysyl-[protein] + 3 S-adenosyl-L-homocysteine
N-terminal L-prolyl-L-prolyl-L-lysyl-[protein] + 2 S-adenosyl-L-methionine = 2 H(+) + N-terminal N,N-dimethyl-L-prolyl-L-prolyl-L-lysyl-[protein] + 2 S-adenosyl-L-homocysteine
N-terminal L-seryl-L-prolyl-L-lysyl-[protein] + 3 S-adenosyl-L-methionine = 3 H(+) + N-terminal N,N,N-trimethyl-L-seryl-L-prolyl-L-lysyl-[protein] + 3 S-adenosyl-L-homocysteine
N-terminal L-prolyl-L-prolyl-L-lysyl-[protein] + 2 S-adenosyl-L-methionine = 2 H(+) + N-terminal N,N-dimethyl-L-prolyl-L-prolyl-L-lysyl-[protein] + 2 S-adenosyl-L-homocysteine
Similarity
Belongs to the methyltransferase superfamily. NTM1 family.
Keywords
Complete proteome
Methyltransferase
Reference proteome
S-adenosyl-L-methionine
Transferase
Feature
chain Alpha N-terminal protein methyltransferase 1
Uniprot
H9J8T5
A0A1E1WA95
A0A2W1BUP6
A0A2H1WR58
A0A2A4K427
S4P8U4
+ More
A0A194RD15 A0A212ETT5 I4DK75 A0A212EHU7 A0A2H1VP09 A0A0L7LVN7 A0A0P4VMP0 R4G383 A0A069DQY0 A0A0L7R794 A0A194QTT4 A0A0N1IQI3 A0A224XS88 A0A088A765 A0A194PMA8 A0A2A3E8E0 A0A154PC33 R4UML6 A0A2J7QPD8 A0A158NPN6 A0A195EL72 E2C5N1 A0A195ETI3 E2AC78 F4W7L1 A0A151WJ87 A0A151IDP6 A0A195ATR5 A0A182FBL3 D6WJE4 A0A182YE73 A0A182SG37 A0A182R8X0 A0A0C9R7A5 A0A026WTT6 A0A067R9C8 A0A182M1H5 E0VEK6 A0A182W8C1 K7ITS6 A0A1S4H3V6 A0A0M8ZST1 Q5TUE6 A0A2P8Z9P4 B3MEZ5 A0A1W4XGU2 A0A084W6U7 R4WDX2 A0A232FBV5 W5JUA1 Q17N31 A0A2M4AYU3 B3N6U6 A0A1B6CF25 B4NWM3 B4HM91 W8C5I1 A0A384ZCM4 Q6NN40 A0A1S4EX08 B4QHS7 A0A1I8PMU0 A0A0K8SZC3 A0A1W4UYZ3 A0A0A9Z911 A0A1B6IWF2 A0A1S3HJA8 A0A182IX10 A0A1S3HJB4 B4KMF7 A0A023ELG9 A0A1B6GQ92 A0A1B6L439 A0A182GF83 A0A3B0JIQ3 B4LNV6 A0A1Q3FJM2 B4MQY2 K1QC85 B4J614 U5EMU9 A0A0T6B7E7 A0A2H8TWN3 A0A0K8UTG4 A0A1A9W3Q4 A0A2R5LG95 A0A034WVW3 C4WXV9 A0A293M4J5 B4GAK0 C1BQM0 A0A1Y1M2K0 A0A182QGN4 Q291L3
A0A194RD15 A0A212ETT5 I4DK75 A0A212EHU7 A0A2H1VP09 A0A0L7LVN7 A0A0P4VMP0 R4G383 A0A069DQY0 A0A0L7R794 A0A194QTT4 A0A0N1IQI3 A0A224XS88 A0A088A765 A0A194PMA8 A0A2A3E8E0 A0A154PC33 R4UML6 A0A2J7QPD8 A0A158NPN6 A0A195EL72 E2C5N1 A0A195ETI3 E2AC78 F4W7L1 A0A151WJ87 A0A151IDP6 A0A195ATR5 A0A182FBL3 D6WJE4 A0A182YE73 A0A182SG37 A0A182R8X0 A0A0C9R7A5 A0A026WTT6 A0A067R9C8 A0A182M1H5 E0VEK6 A0A182W8C1 K7ITS6 A0A1S4H3V6 A0A0M8ZST1 Q5TUE6 A0A2P8Z9P4 B3MEZ5 A0A1W4XGU2 A0A084W6U7 R4WDX2 A0A232FBV5 W5JUA1 Q17N31 A0A2M4AYU3 B3N6U6 A0A1B6CF25 B4NWM3 B4HM91 W8C5I1 A0A384ZCM4 Q6NN40 A0A1S4EX08 B4QHS7 A0A1I8PMU0 A0A0K8SZC3 A0A1W4UYZ3 A0A0A9Z911 A0A1B6IWF2 A0A1S3HJA8 A0A182IX10 A0A1S3HJB4 B4KMF7 A0A023ELG9 A0A1B6GQ92 A0A1B6L439 A0A182GF83 A0A3B0JIQ3 B4LNV6 A0A1Q3FJM2 B4MQY2 K1QC85 B4J614 U5EMU9 A0A0T6B7E7 A0A2H8TWN3 A0A0K8UTG4 A0A1A9W3Q4 A0A2R5LG95 A0A034WVW3 C4WXV9 A0A293M4J5 B4GAK0 C1BQM0 A0A1Y1M2K0 A0A182QGN4 Q291L3
EC Number
2.1.1.244
Pubmed
19121390
28756777
23622113
26354079
22118469
22651552
+ More
26227816 27129103 26334808 21347285 20798317 21719571 18362917 19820115 25244985 24508170 30249741 24845553 20566863 20075255 12364791 29403074 17994087 24438588 23691247 28648823 20920257 23761445 17510324 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 25401762 26823975 24945155 26483478 22992520 25348373 28004739 15632085
26227816 27129103 26334808 21347285 20798317 21719571 18362917 19820115 25244985 24508170 30249741 24845553 20566863 20075255 12364791 29403074 17994087 24438588 23691247 28648823 20920257 23761445 17510324 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 25401762 26823975 24945155 26483478 22992520 25348373 28004739 15632085
EMBL
BABH01005080
GDQN01007189
JAT83865.1
KZ149895
PZC78802.1
ODYU01010437
+ More
SOQ55543.1 NWSH01000172 PCG78776.1 GAIX01009325 JAA83235.1 KQ460398 KPJ15165.1 AGBW02012530 OWR44903.1 AK401693 KQ459601 BAM18315.1 KPI93847.1 AGBW02014796 OWR41059.1 ODYU01003586 SOQ42536.1 JTDY01000006 KOB79500.1 GDKW01002469 JAI54126.1 ACPB03010444 GAHY01001911 JAA75599.1 GBGD01002773 JAC86116.1 KQ414642 KOC66752.1 KQ461154 KPJ08410.1 LADJ01015901 KPJ21027.1 GFTR01003738 JAW12688.1 KQ459599 KPI94457.1 KZ288353 PBC27321.1 KQ434869 KZC09347.1 KC571866 AGM32365.1 NEVH01012093 PNF30439.1 ADTU01022666 KQ978730 KYN28973.1 GL452776 EFN76742.1 KQ981986 KYN31217.1 GL438428 EFN68972.1 GL887844 EGI69884.1 KQ983049 KYQ47870.1 KQ977926 KYM98792.1 KQ976745 KYM75405.1 KQ971342 EFA03687.1 GBYB01012204 JAG81971.1 KK107105 QOIP01000011 EZA59475.1 RLU16422.1 KK852846 KDR15110.1 AXCM01001621 DS235093 EEB11812.1 AAAB01008849 KQ435916 KOX68799.1 EAL41027.3 PYGN01000135 PSN53220.1 CH902619 EDV36616.1 ATLV01020956 KE525310 KFB45941.1 AK417944 BAN21159.1 NNAY01000473 OXU28135.1 ADMH02000071 ETN67907.1 CH477202 EAT48112.1 GGFK01012626 MBW45947.1 CH954177 EDV58195.1 GEDC01025244 JAS12054.1 CM000157 EDW89568.1 CH480816 EDW47168.1 GAMC01001737 JAC04819.1 AE013599 AWM95304.1 BT011456 CM000362 CM002911 EDX06411.1 KMY92598.1 GBRD01007183 JAG58638.1 GBHO01002685 GDHC01006113 JAG40919.1 JAQ12516.1 GECU01016451 JAS91255.1 CH933808 EDW09845.1 GAPW01003500 JAC10098.1 GECZ01005178 JAS64591.1 GEBQ01021586 JAT18391.1 JXUM01059550 KQ562059 KXJ76791.1 OUUW01000001 SPP73309.1 CH940648 EDW61125.2 GFDL01007343 JAV27702.1 CH963849 EDW74521.1 JH818976 EKC26415.1 CH916367 EDW01872.1 GANO01000856 JAB59015.1 LJIG01009341 KRT83274.1 GFXV01006147 MBW17952.1 GDHF01030958 GDHF01022526 JAI21356.1 JAI29788.1 GGLE01004408 MBY08534.1 GAKP01000228 JAC58724.1 ABLF02041191 AK342871 BAH72729.1 GFWV01011164 GFWV01020995 MAA35893.1 CH479181 EDW31952.1 BT076899 ACO11323.1 GEZM01041969 JAV80084.1 AXCN02002298 CM000071 EAL25099.3
SOQ55543.1 NWSH01000172 PCG78776.1 GAIX01009325 JAA83235.1 KQ460398 KPJ15165.1 AGBW02012530 OWR44903.1 AK401693 KQ459601 BAM18315.1 KPI93847.1 AGBW02014796 OWR41059.1 ODYU01003586 SOQ42536.1 JTDY01000006 KOB79500.1 GDKW01002469 JAI54126.1 ACPB03010444 GAHY01001911 JAA75599.1 GBGD01002773 JAC86116.1 KQ414642 KOC66752.1 KQ461154 KPJ08410.1 LADJ01015901 KPJ21027.1 GFTR01003738 JAW12688.1 KQ459599 KPI94457.1 KZ288353 PBC27321.1 KQ434869 KZC09347.1 KC571866 AGM32365.1 NEVH01012093 PNF30439.1 ADTU01022666 KQ978730 KYN28973.1 GL452776 EFN76742.1 KQ981986 KYN31217.1 GL438428 EFN68972.1 GL887844 EGI69884.1 KQ983049 KYQ47870.1 KQ977926 KYM98792.1 KQ976745 KYM75405.1 KQ971342 EFA03687.1 GBYB01012204 JAG81971.1 KK107105 QOIP01000011 EZA59475.1 RLU16422.1 KK852846 KDR15110.1 AXCM01001621 DS235093 EEB11812.1 AAAB01008849 KQ435916 KOX68799.1 EAL41027.3 PYGN01000135 PSN53220.1 CH902619 EDV36616.1 ATLV01020956 KE525310 KFB45941.1 AK417944 BAN21159.1 NNAY01000473 OXU28135.1 ADMH02000071 ETN67907.1 CH477202 EAT48112.1 GGFK01012626 MBW45947.1 CH954177 EDV58195.1 GEDC01025244 JAS12054.1 CM000157 EDW89568.1 CH480816 EDW47168.1 GAMC01001737 JAC04819.1 AE013599 AWM95304.1 BT011456 CM000362 CM002911 EDX06411.1 KMY92598.1 GBRD01007183 JAG58638.1 GBHO01002685 GDHC01006113 JAG40919.1 JAQ12516.1 GECU01016451 JAS91255.1 CH933808 EDW09845.1 GAPW01003500 JAC10098.1 GECZ01005178 JAS64591.1 GEBQ01021586 JAT18391.1 JXUM01059550 KQ562059 KXJ76791.1 OUUW01000001 SPP73309.1 CH940648 EDW61125.2 GFDL01007343 JAV27702.1 CH963849 EDW74521.1 JH818976 EKC26415.1 CH916367 EDW01872.1 GANO01000856 JAB59015.1 LJIG01009341 KRT83274.1 GFXV01006147 MBW17952.1 GDHF01030958 GDHF01022526 JAI21356.1 JAI29788.1 GGLE01004408 MBY08534.1 GAKP01000228 JAC58724.1 ABLF02041191 AK342871 BAH72729.1 GFWV01011164 GFWV01020995 MAA35893.1 CH479181 EDW31952.1 BT076899 ACO11323.1 GEZM01041969 JAV80084.1 AXCN02002298 CM000071 EAL25099.3
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000015103 UP000053825 UP000005203 UP000242457 UP000076502 UP000235965 UP000005205 UP000078492 UP000008237 UP000078541 UP000000311 UP000007755 UP000075809 UP000078542 UP000078540 UP000069272 UP000007266 UP000076408 UP000075901 UP000075900 UP000053097 UP000279307 UP000027135 UP000075883 UP000009046 UP000075920 UP000002358 UP000053105 UP000007062 UP000245037 UP000007801 UP000192223 UP000030765 UP000215335 UP000000673 UP000008820 UP000008711 UP000002282 UP000001292 UP000000803 UP000000304 UP000095300 UP000192221 UP000085678 UP000075880 UP000009192 UP000069940 UP000249989 UP000268350 UP000008792 UP000007798 UP000005408 UP000001070 UP000091820 UP000007819 UP000008744 UP000075886 UP000001819
UP000015103 UP000053825 UP000005203 UP000242457 UP000076502 UP000235965 UP000005205 UP000078492 UP000008237 UP000078541 UP000000311 UP000007755 UP000075809 UP000078542 UP000078540 UP000069272 UP000007266 UP000076408 UP000075901 UP000075900 UP000053097 UP000279307 UP000027135 UP000075883 UP000009046 UP000075920 UP000002358 UP000053105 UP000007062 UP000245037 UP000007801 UP000192223 UP000030765 UP000215335 UP000000673 UP000008820 UP000008711 UP000002282 UP000001292 UP000000803 UP000000304 UP000095300 UP000192221 UP000085678 UP000075880 UP000009192 UP000069940 UP000249989 UP000268350 UP000008792 UP000007798 UP000005408 UP000001070 UP000091820 UP000007819 UP000008744 UP000075886 UP000001819
Pfam
PF05891 Methyltransf_PK
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9J8T5
A0A1E1WA95
A0A2W1BUP6
A0A2H1WR58
A0A2A4K427
S4P8U4
+ More
A0A194RD15 A0A212ETT5 I4DK75 A0A212EHU7 A0A2H1VP09 A0A0L7LVN7 A0A0P4VMP0 R4G383 A0A069DQY0 A0A0L7R794 A0A194QTT4 A0A0N1IQI3 A0A224XS88 A0A088A765 A0A194PMA8 A0A2A3E8E0 A0A154PC33 R4UML6 A0A2J7QPD8 A0A158NPN6 A0A195EL72 E2C5N1 A0A195ETI3 E2AC78 F4W7L1 A0A151WJ87 A0A151IDP6 A0A195ATR5 A0A182FBL3 D6WJE4 A0A182YE73 A0A182SG37 A0A182R8X0 A0A0C9R7A5 A0A026WTT6 A0A067R9C8 A0A182M1H5 E0VEK6 A0A182W8C1 K7ITS6 A0A1S4H3V6 A0A0M8ZST1 Q5TUE6 A0A2P8Z9P4 B3MEZ5 A0A1W4XGU2 A0A084W6U7 R4WDX2 A0A232FBV5 W5JUA1 Q17N31 A0A2M4AYU3 B3N6U6 A0A1B6CF25 B4NWM3 B4HM91 W8C5I1 A0A384ZCM4 Q6NN40 A0A1S4EX08 B4QHS7 A0A1I8PMU0 A0A0K8SZC3 A0A1W4UYZ3 A0A0A9Z911 A0A1B6IWF2 A0A1S3HJA8 A0A182IX10 A0A1S3HJB4 B4KMF7 A0A023ELG9 A0A1B6GQ92 A0A1B6L439 A0A182GF83 A0A3B0JIQ3 B4LNV6 A0A1Q3FJM2 B4MQY2 K1QC85 B4J614 U5EMU9 A0A0T6B7E7 A0A2H8TWN3 A0A0K8UTG4 A0A1A9W3Q4 A0A2R5LG95 A0A034WVW3 C4WXV9 A0A293M4J5 B4GAK0 C1BQM0 A0A1Y1M2K0 A0A182QGN4 Q291L3
A0A194RD15 A0A212ETT5 I4DK75 A0A212EHU7 A0A2H1VP09 A0A0L7LVN7 A0A0P4VMP0 R4G383 A0A069DQY0 A0A0L7R794 A0A194QTT4 A0A0N1IQI3 A0A224XS88 A0A088A765 A0A194PMA8 A0A2A3E8E0 A0A154PC33 R4UML6 A0A2J7QPD8 A0A158NPN6 A0A195EL72 E2C5N1 A0A195ETI3 E2AC78 F4W7L1 A0A151WJ87 A0A151IDP6 A0A195ATR5 A0A182FBL3 D6WJE4 A0A182YE73 A0A182SG37 A0A182R8X0 A0A0C9R7A5 A0A026WTT6 A0A067R9C8 A0A182M1H5 E0VEK6 A0A182W8C1 K7ITS6 A0A1S4H3V6 A0A0M8ZST1 Q5TUE6 A0A2P8Z9P4 B3MEZ5 A0A1W4XGU2 A0A084W6U7 R4WDX2 A0A232FBV5 W5JUA1 Q17N31 A0A2M4AYU3 B3N6U6 A0A1B6CF25 B4NWM3 B4HM91 W8C5I1 A0A384ZCM4 Q6NN40 A0A1S4EX08 B4QHS7 A0A1I8PMU0 A0A0K8SZC3 A0A1W4UYZ3 A0A0A9Z911 A0A1B6IWF2 A0A1S3HJA8 A0A182IX10 A0A1S3HJB4 B4KMF7 A0A023ELG9 A0A1B6GQ92 A0A1B6L439 A0A182GF83 A0A3B0JIQ3 B4LNV6 A0A1Q3FJM2 B4MQY2 K1QC85 B4J614 U5EMU9 A0A0T6B7E7 A0A2H8TWN3 A0A0K8UTG4 A0A1A9W3Q4 A0A2R5LG95 A0A034WVW3 C4WXV9 A0A293M4J5 B4GAK0 C1BQM0 A0A1Y1M2K0 A0A182QGN4 Q291L3
PDB
5CVE
E-value=2.80478e-49,
Score=490
Ontologies
GO
PANTHER
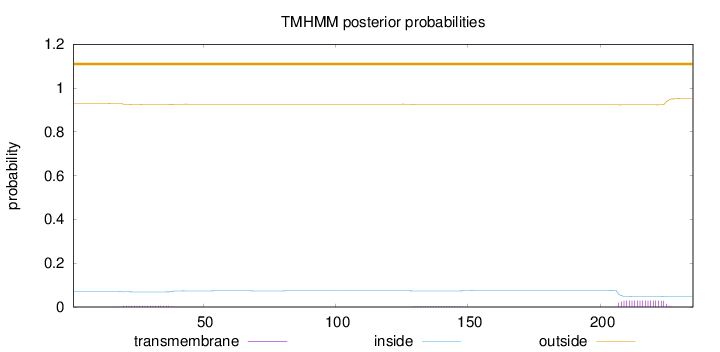
Topology
Length:
235
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.6954
Exp number, first 60 AAs:
0.10987
Total prob of N-in:
0.07011
outside
1 - 235
Population Genetic Test Statistics
Pi
257.722356
Theta
204.776181
Tajima's D
0.469729
CLR
0
CSRT
0.504124793760312
Interpretation
Uncertain
