Gene
KWMTBOMO01940
Pre Gene Modal
BGIBMGA005929
Annotation
PREDICTED:_protein-O-fucosyltransferase_1_isoform_X1_[Bombyx_mori]
Full name
GDP-fucose protein O-fucosyltransferase 1
Alternative Name
Neurotic protein
Peptide-O-fucosyltransferase 1
Peptide-O-fucosyltransferase 1
Location in the cell
Cytoplasmic Reliability : 2.417 Nuclear Reliability : 2.133
Sequence
CDS
ATGGGCAGATTTGGGAACCAAGCTGATCACTTTCTGGGTGTTTTGGCTTTTAGTAAAGCAGTTAATAGAACTTTAATTCTGCCACCTTGGATTGAATACAGTATGGCCGATAGAAAAGAGAGTCTCTTTTTGCTACACTCAAAGGACAGATCAGAGTTTTATGCACCTTTGAATTATGATGCTCAGAATGAAAAGATCTTGGAACAGTGGAAAATAAGGTATCCAGTTGATAAATGGCCTGTTTTAGCCTTTACTGGTGCTCCAGCTAGTTTTCCAGTACAAAAAGAGAATGTGGATATGCAGAAATATTTAAAATGGAGTAATGATATTGAACACAGATCAAAACTTTTTATAAAAAAGAACATGAGTGGAGGAGGTTTTCTTGGAATTCATCTCAGAAATGGACAAGACTGGGTAAAAGCATGCCAGCATGTAAAAGACAGTCCAATGCTGTTTGCAGCACCACAATGCGTTGGCTACAGAAATGAACGAGGCCCATTAACAATGTCAATGTGTTTACCAAAGCCTTCTGAAATCTTAAAGCAAATAAAACGAGTAATAAAGAAACTTAGCGACATTAAATATATTTTTGTGGCGTCCGACTCTAACCATATGATAGAAGAATTAAATGCTGGCCTGCGTCGTCCAGAAATAAAGATTTTAAAATTTCAGCCATCGTCCAATCCACACTTGGATTTAGCTATTCTTGGACAAGCAAATTATTTTATTGGAAATTGTGTATCGTCATATTCAGCATTTGTCAAAAGGGAACGTGACATGAAAGGGTTACCTTCAGAATTTTGGTCATTTCCACAACGAAAGAAAAACAAACATGAAGAACTATAG
Protein
MGRFGNQADHFLGVLAFSKAVNRTLILPPWIEYSMADRKESLFLLHSKDRSEFYAPLNYDAQNEKILEQWKIRYPVDKWPVLAFTGAPASFPVQKENVDMQKYLKWSNDIEHRSKLFIKKNMSGGGFLGIHLRNGQDWVKACQHVKDSPMLFAAPQCVGYRNERGPLTMSMCLPKPSEILKQIKRVIKKLSDIKYIFVASDSNHMIEELNAGLRRPEIKILKFQPSSNPHLDLAILGQANYFIGNCVSSYSAFVKRERDMKGLPSEFWSFPQRKKNKHEEL
Summary
Description
Catalyzes the reaction that attaches fucose through an O-glycosidic linkage to a conserved serine or threonine residue found in the consensus sequence C2-X(4,5)-[S/T]-C3 of EGF domains, where C2 and C3 are the second and third conserved cysteines. Specifically uses GDP-fucose as donor substrate and proper disulfide pairing of the substrate EGF domains is required for fucose transfer. Plays a crucial role in Notch signaling. Initial fucosylation of Notch/N by POFUT1 generates a substrate for Fringe/Fng, an acetylglucosaminyltransferase that can then extend the fucosylation on the Notch EGF repeats. This extended fucosylation is required for optimal ligand binding and canonical NOTCH signaling induced by Delta/Dl or Serrate/Ser. Also required for the transport of Notch/N to the early endosome.
Catalytic Activity
Transfers an alpha-L-fucosyl residue from GDP-beta-L-fucose to the serine hydroxy group of a protein acceptor.
Subunit
Interacts with N (via its EGF domains); the interaction is required for the endocyctic transport of N.
Similarity
Belongs to the glycosyltransferase 65 family.
Belongs to the RecA family.
Belongs to the RecA family.
Keywords
Carbohydrate metabolism
Complete proteome
Disulfide bond
Endoplasmic reticulum
Fucose metabolism
Glycoprotein
Glycosyltransferase
Notch signaling pathway
Reference proteome
Signal
Transferase
Feature
chain GDP-fucose protein O-fucosyltransferase 1
Uniprot
H9J8T7
Q659S0
Q00P42
A0A2H1WR48
A0A2W1BV04
A0A0L7LML0
+ More
A0A194RD10 A0A212ETX2 A0A194PLH0 S4PPE1 A0A2A4K4F0 A0A1B6ILL0 A0A0J7NKK3 E1ZV69 A0A1I8MTV4 A0A1B6M642 D6WLU5 A0A2A3ENF8 B3NRC7 A0A1W4WKV2 A0A067RR47 A0A1Q3FHL3 A0A1Q3FHN2 B4JW84 B4QFD1 A0A0M8ZUH9 E0VCP3 B0X217 A0A0C9QJG1 Q659S1 A0A1Q3FH50 A0A1Q3FH11 E2BD92 K7J3H8 A0A182Y708 A8E6L6 Q9V6X7 A0A154PEI3 A0A232FEA8 A0A034WMV2 A0A1B6CCN0 B3MCZ6 A0A0K8U5B3 U4UHL0 A0A0L0BU95 A0A0M4EVV4 A0A1B6ER18 N6TUE8 B4HR13 A0A182P5P8 B4LJU0 A0A1W4VCK8 A0A0K8TKW5 A0A0L7QZ02 A0A026X319 A0A2S2PAA8 A0A182N3C1 J9KAW3 Q6EV68 Q292F1 A0A1I8NLW7 A0A293LZ27 B4GDM4 A0A182MA77 A0A0A1WL84 B4KT64 A0A1Y1MQ91 A0A182QNI9 A0A084WCI7 Q00P44 A0A2H8TGU7 Q17N26 A0A3B0JGA0 A0A336LRQ0 A0A1B0CRL3 A0A2J7RF22 A0A1A9Z1W3 B4MY74 A0A336MZV8 A0A336MRK8 A0A1J1IZC1 A0A1A9V1L9 A0A1A9W3R1 W8BMH7 V5GV70 Q7QHS7 A0A182LH93 A0A182JU40 A0A182HDQ7 A0A182HY47 A0A182WXH2 T1H0K8 A0A195FGC2 A0A151XAH6 A0A182VAD0 A0A182RLI8 A0A1B0D685 A0A182TPG4 A0A1B0F9G3 A0A158NGP3
A0A194RD10 A0A212ETX2 A0A194PLH0 S4PPE1 A0A2A4K4F0 A0A1B6ILL0 A0A0J7NKK3 E1ZV69 A0A1I8MTV4 A0A1B6M642 D6WLU5 A0A2A3ENF8 B3NRC7 A0A1W4WKV2 A0A067RR47 A0A1Q3FHL3 A0A1Q3FHN2 B4JW84 B4QFD1 A0A0M8ZUH9 E0VCP3 B0X217 A0A0C9QJG1 Q659S1 A0A1Q3FH50 A0A1Q3FH11 E2BD92 K7J3H8 A0A182Y708 A8E6L6 Q9V6X7 A0A154PEI3 A0A232FEA8 A0A034WMV2 A0A1B6CCN0 B3MCZ6 A0A0K8U5B3 U4UHL0 A0A0L0BU95 A0A0M4EVV4 A0A1B6ER18 N6TUE8 B4HR13 A0A182P5P8 B4LJU0 A0A1W4VCK8 A0A0K8TKW5 A0A0L7QZ02 A0A026X319 A0A2S2PAA8 A0A182N3C1 J9KAW3 Q6EV68 Q292F1 A0A1I8NLW7 A0A293LZ27 B4GDM4 A0A182MA77 A0A0A1WL84 B4KT64 A0A1Y1MQ91 A0A182QNI9 A0A084WCI7 Q00P44 A0A2H8TGU7 Q17N26 A0A3B0JGA0 A0A336LRQ0 A0A1B0CRL3 A0A2J7RF22 A0A1A9Z1W3 B4MY74 A0A336MZV8 A0A336MRK8 A0A1J1IZC1 A0A1A9V1L9 A0A1A9W3R1 W8BMH7 V5GV70 Q7QHS7 A0A182LH93 A0A182JU40 A0A182HDQ7 A0A182HY47 A0A182WXH2 T1H0K8 A0A195FGC2 A0A151XAH6 A0A182VAD0 A0A182RLI8 A0A1B0D685 A0A182TPG4 A0A1B0F9G3 A0A158NGP3
EC Number
2.4.1.221
Pubmed
19121390
16679357
28756777
26227816
26354079
22118469
+ More
23622113 20798317 25315136 18362917 19820115 17994087 24845553 22936249 20566863 17550304 20075255 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12917292 12537569 11524432 11698403 12909620 17329366 28648823 25348373 23537049 26108605 26369729 24508170 30249741 12966037 15632085 23185243 25830018 28004739 24438588 17510324 24495485 12364791 14747013 17210077 20966253 26483478 21347285
23622113 20798317 25315136 18362917 19820115 17994087 24845553 22936249 20566863 17550304 20075255 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12917292 12537569 11524432 11698403 12909620 17329366 28648823 25348373 23537049 26108605 26369729 24508170 30249741 12966037 15632085 23185243 25830018 28004739 24438588 17510324 24495485 12364791 14747013 17210077 20966253 26483478 21347285
EMBL
BABH01005075
BABH01005076
AJ831491
CAH40835.1
DQ139946
ABA29467.1
+ More
ODYU01010437 SOQ55545.1 KZ149895 PZC78799.1 JTDY01000569 KOB76645.1 KQ460398 KPJ15160.1 AGBW02012530 OWR44901.1 KQ459601 KPI93843.1 GAIX01000302 JAA92258.1 NWSH01000172 PCG78788.1 GECU01019895 JAS87811.1 LBMM01003913 KMQ93000.1 GL434441 EFN74928.1 GEBQ01008577 JAT31400.1 KQ971343 EFA03399.2 KZ288204 PBC33230.1 CH954179 EDV56079.1 KK852514 KDR22209.1 GFDL01008007 JAV27038.1 GFDL01007987 JAV27058.1 CH916375 EDV98222.1 CM000362 CM002911 EDX07041.1 KMY93690.1 KQ435840 KOX71405.1 DS235060 EEB11149.1 DS232276 EDS38947.1 GBYB01000702 JAG70469.1 AJ831490 CM000158 CAH40834.1 EDW90977.1 GFDL01008186 JAV26859.1 GFDL01008185 JAV26860.1 GL447585 EFN86320.1 BT030808 AE013599 ABV82190.1 AHN56204.1 AB093572 BT021295 AY118651 KQ434874 KZC09688.1 NNAY01000337 OXU29106.1 GAKP01002066 JAC56886.1 GEDC01026119 GEDC01008701 JAS11179.1 JAS28597.1 CH902619 EDV36311.1 GDHF01030462 JAI21852.1 KB632327 ERL92517.1 JRES01001317 KNC23650.1 CP012524 ALC41820.1 GECZ01029368 JAS40401.1 APGK01051866 KB741207 ENN72900.1 CH480816 EDW47810.1 CH940648 EDW60599.2 GDAI01002629 JAI14974.1 KQ414685 KOC63801.1 KK107020 QOIP01000004 EZA62421.1 RLU23223.1 GGMR01013756 MBY26375.1 ABLF02039399 AJ781501 CAH03716.1 DQ139947 CM000071 ABA29468.1 EAL24911.1 GFWV01008372 MAA33101.1 CH479181 EDW31681.1 AXCM01001256 GBXI01015022 JAC99269.1 CH933808 EDW09584.2 GEZM01024260 JAV87822.1 AXCN02001113 ATLV01022672 KE525335 KFB47931.1 DQ139944 ABA29465.1 GFXV01001519 MBW13324.1 CH477202 EAT48119.1 OUUW01000001 SPP74380.1 UFQS01004263 UFQT01004263 SSX16292.1 SSX35612.1 AJWK01024922 NEVH01004414 PNF39432.1 CH963894 EDW77063.1 UFQT01005066 SSX35882.1 UFQS01002304 UFQT01002304 SSX13753.1 SSX33174.1 CVRI01000064 CRL05440.1 GAMC01008372 JAB98183.1 GALX01000432 JAB68034.1 DQ139945 AAAB01008816 ABA29466.1 EAA05278.4 JXUM01130315 JXUM01130316 KQ567562 KXJ69367.1 APCN01004491 CAQQ02373491 KQ981625 KYN39064.1 KQ982339 KYQ57339.1 AJVK01012008 CCAG010002095 ADTU01015163 ADTU01015164
ODYU01010437 SOQ55545.1 KZ149895 PZC78799.1 JTDY01000569 KOB76645.1 KQ460398 KPJ15160.1 AGBW02012530 OWR44901.1 KQ459601 KPI93843.1 GAIX01000302 JAA92258.1 NWSH01000172 PCG78788.1 GECU01019895 JAS87811.1 LBMM01003913 KMQ93000.1 GL434441 EFN74928.1 GEBQ01008577 JAT31400.1 KQ971343 EFA03399.2 KZ288204 PBC33230.1 CH954179 EDV56079.1 KK852514 KDR22209.1 GFDL01008007 JAV27038.1 GFDL01007987 JAV27058.1 CH916375 EDV98222.1 CM000362 CM002911 EDX07041.1 KMY93690.1 KQ435840 KOX71405.1 DS235060 EEB11149.1 DS232276 EDS38947.1 GBYB01000702 JAG70469.1 AJ831490 CM000158 CAH40834.1 EDW90977.1 GFDL01008186 JAV26859.1 GFDL01008185 JAV26860.1 GL447585 EFN86320.1 BT030808 AE013599 ABV82190.1 AHN56204.1 AB093572 BT021295 AY118651 KQ434874 KZC09688.1 NNAY01000337 OXU29106.1 GAKP01002066 JAC56886.1 GEDC01026119 GEDC01008701 JAS11179.1 JAS28597.1 CH902619 EDV36311.1 GDHF01030462 JAI21852.1 KB632327 ERL92517.1 JRES01001317 KNC23650.1 CP012524 ALC41820.1 GECZ01029368 JAS40401.1 APGK01051866 KB741207 ENN72900.1 CH480816 EDW47810.1 CH940648 EDW60599.2 GDAI01002629 JAI14974.1 KQ414685 KOC63801.1 KK107020 QOIP01000004 EZA62421.1 RLU23223.1 GGMR01013756 MBY26375.1 ABLF02039399 AJ781501 CAH03716.1 DQ139947 CM000071 ABA29468.1 EAL24911.1 GFWV01008372 MAA33101.1 CH479181 EDW31681.1 AXCM01001256 GBXI01015022 JAC99269.1 CH933808 EDW09584.2 GEZM01024260 JAV87822.1 AXCN02001113 ATLV01022672 KE525335 KFB47931.1 DQ139944 ABA29465.1 GFXV01001519 MBW13324.1 CH477202 EAT48119.1 OUUW01000001 SPP74380.1 UFQS01004263 UFQT01004263 SSX16292.1 SSX35612.1 AJWK01024922 NEVH01004414 PNF39432.1 CH963894 EDW77063.1 UFQT01005066 SSX35882.1 UFQS01002304 UFQT01002304 SSX13753.1 SSX33174.1 CVRI01000064 CRL05440.1 GAMC01008372 JAB98183.1 GALX01000432 JAB68034.1 DQ139945 AAAB01008816 ABA29466.1 EAA05278.4 JXUM01130315 JXUM01130316 KQ567562 KXJ69367.1 APCN01004491 CAQQ02373491 KQ981625 KYN39064.1 KQ982339 KYQ57339.1 AJVK01012008 CCAG010002095 ADTU01015163 ADTU01015164
Proteomes
UP000005204
UP000037510
UP000053240
UP000007151
UP000053268
UP000218220
+ More
UP000036403 UP000000311 UP000095301 UP000007266 UP000242457 UP000008711 UP000192223 UP000027135 UP000001070 UP000000304 UP000053105 UP000009046 UP000002320 UP000002282 UP000008237 UP000002358 UP000076408 UP000000803 UP000076502 UP000215335 UP000007801 UP000030742 UP000037069 UP000092553 UP000019118 UP000001292 UP000075885 UP000008792 UP000192221 UP000053825 UP000053097 UP000279307 UP000075884 UP000007819 UP000001819 UP000095300 UP000008744 UP000075883 UP000009192 UP000075886 UP000030765 UP000008820 UP000268350 UP000092461 UP000235965 UP000092445 UP000007798 UP000183832 UP000078200 UP000091820 UP000007062 UP000075882 UP000075881 UP000069940 UP000249989 UP000075840 UP000076407 UP000015102 UP000078541 UP000075809 UP000075903 UP000075900 UP000092462 UP000075902 UP000092444 UP000005205
UP000036403 UP000000311 UP000095301 UP000007266 UP000242457 UP000008711 UP000192223 UP000027135 UP000001070 UP000000304 UP000053105 UP000009046 UP000002320 UP000002282 UP000008237 UP000002358 UP000076408 UP000000803 UP000076502 UP000215335 UP000007801 UP000030742 UP000037069 UP000092553 UP000019118 UP000001292 UP000075885 UP000008792 UP000192221 UP000053825 UP000053097 UP000279307 UP000075884 UP000007819 UP000001819 UP000095300 UP000008744 UP000075883 UP000009192 UP000075886 UP000030765 UP000008820 UP000268350 UP000092461 UP000235965 UP000092445 UP000007798 UP000183832 UP000078200 UP000091820 UP000007062 UP000075882 UP000075881 UP000069940 UP000249989 UP000075840 UP000076407 UP000015102 UP000078541 UP000075809 UP000075903 UP000075900 UP000092462 UP000075902 UP000092444 UP000005205
Interpro
ProteinModelPortal
H9J8T7
Q659S0
Q00P42
A0A2H1WR48
A0A2W1BV04
A0A0L7LML0
+ More
A0A194RD10 A0A212ETX2 A0A194PLH0 S4PPE1 A0A2A4K4F0 A0A1B6ILL0 A0A0J7NKK3 E1ZV69 A0A1I8MTV4 A0A1B6M642 D6WLU5 A0A2A3ENF8 B3NRC7 A0A1W4WKV2 A0A067RR47 A0A1Q3FHL3 A0A1Q3FHN2 B4JW84 B4QFD1 A0A0M8ZUH9 E0VCP3 B0X217 A0A0C9QJG1 Q659S1 A0A1Q3FH50 A0A1Q3FH11 E2BD92 K7J3H8 A0A182Y708 A8E6L6 Q9V6X7 A0A154PEI3 A0A232FEA8 A0A034WMV2 A0A1B6CCN0 B3MCZ6 A0A0K8U5B3 U4UHL0 A0A0L0BU95 A0A0M4EVV4 A0A1B6ER18 N6TUE8 B4HR13 A0A182P5P8 B4LJU0 A0A1W4VCK8 A0A0K8TKW5 A0A0L7QZ02 A0A026X319 A0A2S2PAA8 A0A182N3C1 J9KAW3 Q6EV68 Q292F1 A0A1I8NLW7 A0A293LZ27 B4GDM4 A0A182MA77 A0A0A1WL84 B4KT64 A0A1Y1MQ91 A0A182QNI9 A0A084WCI7 Q00P44 A0A2H8TGU7 Q17N26 A0A3B0JGA0 A0A336LRQ0 A0A1B0CRL3 A0A2J7RF22 A0A1A9Z1W3 B4MY74 A0A336MZV8 A0A336MRK8 A0A1J1IZC1 A0A1A9V1L9 A0A1A9W3R1 W8BMH7 V5GV70 Q7QHS7 A0A182LH93 A0A182JU40 A0A182HDQ7 A0A182HY47 A0A182WXH2 T1H0K8 A0A195FGC2 A0A151XAH6 A0A182VAD0 A0A182RLI8 A0A1B0D685 A0A182TPG4 A0A1B0F9G3 A0A158NGP3
A0A194RD10 A0A212ETX2 A0A194PLH0 S4PPE1 A0A2A4K4F0 A0A1B6ILL0 A0A0J7NKK3 E1ZV69 A0A1I8MTV4 A0A1B6M642 D6WLU5 A0A2A3ENF8 B3NRC7 A0A1W4WKV2 A0A067RR47 A0A1Q3FHL3 A0A1Q3FHN2 B4JW84 B4QFD1 A0A0M8ZUH9 E0VCP3 B0X217 A0A0C9QJG1 Q659S1 A0A1Q3FH50 A0A1Q3FH11 E2BD92 K7J3H8 A0A182Y708 A8E6L6 Q9V6X7 A0A154PEI3 A0A232FEA8 A0A034WMV2 A0A1B6CCN0 B3MCZ6 A0A0K8U5B3 U4UHL0 A0A0L0BU95 A0A0M4EVV4 A0A1B6ER18 N6TUE8 B4HR13 A0A182P5P8 B4LJU0 A0A1W4VCK8 A0A0K8TKW5 A0A0L7QZ02 A0A026X319 A0A2S2PAA8 A0A182N3C1 J9KAW3 Q6EV68 Q292F1 A0A1I8NLW7 A0A293LZ27 B4GDM4 A0A182MA77 A0A0A1WL84 B4KT64 A0A1Y1MQ91 A0A182QNI9 A0A084WCI7 Q00P44 A0A2H8TGU7 Q17N26 A0A3B0JGA0 A0A336LRQ0 A0A1B0CRL3 A0A2J7RF22 A0A1A9Z1W3 B4MY74 A0A336MZV8 A0A336MRK8 A0A1J1IZC1 A0A1A9V1L9 A0A1A9W3R1 W8BMH7 V5GV70 Q7QHS7 A0A182LH93 A0A182JU40 A0A182HDQ7 A0A182HY47 A0A182WXH2 T1H0K8 A0A195FGC2 A0A151XAH6 A0A182VAD0 A0A182RLI8 A0A1B0D685 A0A182TPG4 A0A1B0F9G3 A0A158NGP3
PDB
5KXQ
E-value=9.52524e-42,
Score=426
Ontologies
GO
PANTHER
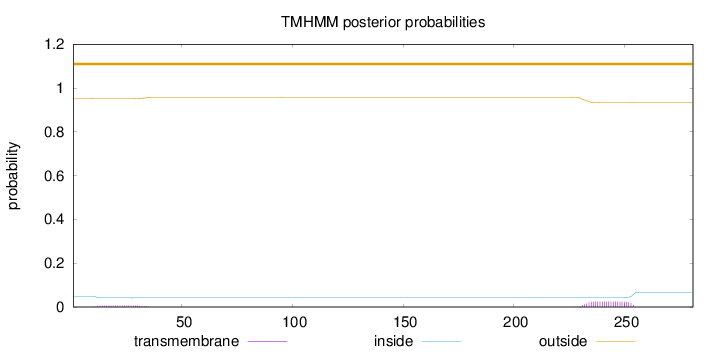
Topology
Subcellular location
Endoplasmic reticulum
Length:
281
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.66774
Exp number, first 60 AAs:
0.144
Total prob of N-in:
0.04793
outside
1 - 281
Population Genetic Test Statistics
Pi
33.709744
Theta
59.529339
Tajima's D
0.105493
CLR
0.29612
CSRT
0.393830308484576
Interpretation
Uncertain
