Pre Gene Modal
BGIBMGA006221
Annotation
PREDICTED:_ras-related_protein_Rab-14_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.151 Mitochondrial Reliability : 1.228
Sequence
CDS
ATGACTTCAGGACCCTATAACTACTCATACATATTTAAGTATATTATAATTGGAGATATGGGAGTTGGTAAATCATGCCTCTTGCATCAATTCACTGATAAAAAATTCATGGCTGATTGTCCTCACACTATTGGTGTTGAGTTTGGAACGCGTATAATCGAAGTTGCTGGACAAAAAATCAAGCTACAGATTTGGGATACAGCTGGACAAGAAAGGTTTCGTGCTGTCACTCGATCTTACTACAGAGGTGCAGCTGGAGCTCTTATGGTTTATGACATAACAAGAAGATCCACATATAATCACCTTAGCAGCTGGCTGACAGATACTCGTAATCTGACTAATCCTAGCACTGTGATATTTTTAATTGGCAATAAATCCGATTTAGATGCCCAAAGAGATGTTACTTATGAGGAAGCTAAACAATTTGCTGATGAAAATGGCTTGATGTTTGTTGAAGCAAGTGCTAAAACAGGGCAGAATGTCGAGGAAGCATTTTTGGAGACCGCCAAAAAAATATACCAGAGCATACAAGATGGCCGGCTGGACTTGAACGCGGCGGAGTCGGGCGTCCAGCACAAGCCCGCGTCCCCGGGCCGCCCGCTCGCCGCGCCGCCCTCCTCGCGCGACAACTGCGCGTGCTAA
Protein
MTSGPYNYSYIFKYIIIGDMGVGKSCLLHQFTDKKFMADCPHTIGVEFGTRIIEVAGQKIKLQIWDTAGQERFRAVTRSYYRGAAGALMVYDITRRSTYNHLSSWLTDTRNLTNPSTVIFLIGNKSDLDAQRDVTYEEAKQFADENGLMFVEASAKTGQNVEEAFLETAKKIYQSIQDGRLDLNAAESGVQHKPASPGRPLAAPPSSRDNCAC
Summary
Uniprot
A0A2W1BUQ7
A0A2H1WYQ7
A0A2A4K2R8
A0A194PRU1
A0A194RC43
A0A212ETT0
+ More
S4P930 A0A067R9C4 A0A1Y1KYF7 A0A232ENP5 A0A0B4UC52 A0A1W4XFX6 A0A0L7RA24 A0A2A3EL95 V9IIX3 A0A0N0BH50 A0A088A3U8 F4W7C1 A0A0J7KXD2 A0A158NPW7 A0A151X8V7 A0A026WIL4 A0A195CJ97 A0A195DSS0 A0A0K8SLD5 A0A154P250 N6U3U2 A0A1L8DV52 A0A1L8DV18 A0A0P4VRG0 R4G4K6 A0A2L2Y4L3 E9IRE0 A0A224XXD2 K7IXC5 A0A164TRN1 V3YVU6 B0WCS3 A0A310SXR9 H2Y4P3 A0A2R7VZ04 A0A1S3HQH2 E9HWB8 A0A1Q3FHB9 A0A0N8CVH9 E7BTP8 A0A023EJU0 Q16JQ5 C3ZTG4 S4RAH7 A0A2C9JWN1 A0A0B7AZA8 Q16JQ4 A0A023EKN9 A0A1Q3FH10 A0A131XU96 K1QC78 R7VJW8 A0A0P6HF76 A0A1S3PRE2 A0A2R5L800 A0A293LTH8 F6VMY2 A0A3P8XXR9 A0A060XNR3 A0A3B3RXK7 A0A2I4CVT3 A0A210PSV6 A0A384CRD0 L8IY39 F7B9P4 S7PXR1 G1N6Z1 A0A3B4XAX4 A0A3B3YHN5 A0A091GZ27 A0A3B5BC83 A0A099ZMY2 A0A151MG19 W5ME44 A0A091IHD3 A0A091GJI0 A0A091URK5 A0A218V634 A0A093PMB1 M3ZSX6 A0A091WL45 K7F802 A0A087VAU9 A0A3B4TIX6 A0A3B3BBX7 A0A1A8GCU5 A0A3B3TS66 H9GDR1 A0A3P8YA38 A0A1A8LI77 A0A0B8RPH2 A0A093CK25 A0A1A8E6A0 A0A091IR10
S4P930 A0A067R9C4 A0A1Y1KYF7 A0A232ENP5 A0A0B4UC52 A0A1W4XFX6 A0A0L7RA24 A0A2A3EL95 V9IIX3 A0A0N0BH50 A0A088A3U8 F4W7C1 A0A0J7KXD2 A0A158NPW7 A0A151X8V7 A0A026WIL4 A0A195CJ97 A0A195DSS0 A0A0K8SLD5 A0A154P250 N6U3U2 A0A1L8DV52 A0A1L8DV18 A0A0P4VRG0 R4G4K6 A0A2L2Y4L3 E9IRE0 A0A224XXD2 K7IXC5 A0A164TRN1 V3YVU6 B0WCS3 A0A310SXR9 H2Y4P3 A0A2R7VZ04 A0A1S3HQH2 E9HWB8 A0A1Q3FHB9 A0A0N8CVH9 E7BTP8 A0A023EJU0 Q16JQ5 C3ZTG4 S4RAH7 A0A2C9JWN1 A0A0B7AZA8 Q16JQ4 A0A023EKN9 A0A1Q3FH10 A0A131XU96 K1QC78 R7VJW8 A0A0P6HF76 A0A1S3PRE2 A0A2R5L800 A0A293LTH8 F6VMY2 A0A3P8XXR9 A0A060XNR3 A0A3B3RXK7 A0A2I4CVT3 A0A210PSV6 A0A384CRD0 L8IY39 F7B9P4 S7PXR1 G1N6Z1 A0A3B4XAX4 A0A3B3YHN5 A0A091GZ27 A0A3B5BC83 A0A099ZMY2 A0A151MG19 W5ME44 A0A091IHD3 A0A091GJI0 A0A091URK5 A0A218V634 A0A093PMB1 M3ZSX6 A0A091WL45 K7F802 A0A087VAU9 A0A3B4TIX6 A0A3B3BBX7 A0A1A8GCU5 A0A3B3TS66 H9GDR1 A0A3P8YA38 A0A1A8LI77 A0A0B8RPH2 A0A093CK25 A0A1A8E6A0 A0A091IR10
Pubmed
28756777
26354079
22118469
23622113
24845553
28004739
+ More
28648823 25542378 21719571 21347285 24508170 30249741 23537049 27129103 26561354 21282665 20075255 23254933 21292972 24945155 17510324 18563158 15562597 26483478 22992520 12481130 15114417 25069045 24755649 29240929 28812685 22751099 19892987 20838655 22293439 23542700 17381049 29451363 21881562 25476704
28648823 25542378 21719571 21347285 24508170 30249741 23537049 27129103 26561354 21282665 20075255 23254933 21292972 24945155 17510324 18563158 15562597 26483478 22992520 12481130 15114417 25069045 24755649 29240929 28812685 22751099 19892987 20838655 22293439 23542700 17381049 29451363 21881562 25476704
EMBL
KZ149895
PZC78812.1
ODYU01012086
SOQ58199.1
NWSH01000193
PCG78545.1
+ More
KQ459601 KPI93855.1 KQ460398 KPJ15172.1 AGBW02012530 OWR44912.1 GAIX01006992 JAA85568.1 KK852846 KDR15105.1 GEZM01075860 GEZM01075858 GEZM01075857 JAV63917.1 NNAY01003108 OXU19958.1 KP216762 AJC97119.1 KQ414621 KOC67606.1 KZ288225 PBC31929.1 JR048118 AEY60582.1 KQ435759 KOX75731.1 GL887844 EGI69794.1 LBMM01002182 KMQ95172.1 ADTU01022791 KQ982409 KYQ56750.1 KK107183 QOIP01000011 EZA55860.1 RLU17110.1 KQ977649 KYN00816.1 KQ980487 KYN15893.1 GBRD01012219 JAG53605.1 KQ434787 KZC05298.1 APGK01040737 APGK01040738 KB740984 KB631904 ENN76260.1 ERL87087.1 GFDF01003785 JAV10299.1 GFDF01003806 JAV10278.1 GDKW01000960 JAI55635.1 ACPB03001109 GAHY01000691 JAA76819.1 IAAA01001821 IAAA01001822 LAA03111.1 GL765107 EFZ16912.1 GFTR01003603 JAW12823.1 AAZX01000032 AAZX01016030 LRGB01001745 KZS10691.1 KB204017 ESO82123.1 DS231890 EDS43799.1 KQ759768 OAD62949.1 KK854094 PTY11165.1 GL732911 EFX63965.1 GFDL01008110 JAV26935.1 GDIP01094815 JAM08900.1 GU289394 ADK66883.1 GAPW01004256 JAC09342.1 CH477997 EAT34504.1 GG666677 EEN44211.1 HACG01038531 CEK85396.1 EAT34505.2 JXUM01131546 GAPW01003845 KQ567724 JAC09753.1 KXJ69305.1 GFDL01008252 JAV26793.1 GEFM01005894 JAP69902.1 JH817502 EKC28779.1 AMQN01004337 KB293237 ELU16260.1 GDIQ01019938 JAN74799.1 GGLE01001486 MBY05612.1 GFWV01006348 MAA31078.1 EAAA01002795 FR905707 CDQ81111.1 NEDP02005521 OWF39565.1 JH880525 ELR60903.1 KE164180 EPQ15773.1 KL517270 KFO88424.1 KL896018 KGL83122.1 AKHW03006215 KYO23451.1 AHAT01024887 KL217813 KFO99130.1 KL448313 KFO82555.1 KL409973 KFQ93042.1 MUZQ01000043 OWK61379.1 KL414671 KFW77973.1 KK735751 KFR15638.1 AGCU01069984 AGCU01069985 KL486402 KFO09741.1 HAEC01000112 SBQ68189.1 HAEF01006930 SBR44312.1 GBSH01002177 JAG66849.1 KL245982 KFV14843.1 HAEA01013013 SBQ41493.1 KK500860 KFP11139.1
KQ459601 KPI93855.1 KQ460398 KPJ15172.1 AGBW02012530 OWR44912.1 GAIX01006992 JAA85568.1 KK852846 KDR15105.1 GEZM01075860 GEZM01075858 GEZM01075857 JAV63917.1 NNAY01003108 OXU19958.1 KP216762 AJC97119.1 KQ414621 KOC67606.1 KZ288225 PBC31929.1 JR048118 AEY60582.1 KQ435759 KOX75731.1 GL887844 EGI69794.1 LBMM01002182 KMQ95172.1 ADTU01022791 KQ982409 KYQ56750.1 KK107183 QOIP01000011 EZA55860.1 RLU17110.1 KQ977649 KYN00816.1 KQ980487 KYN15893.1 GBRD01012219 JAG53605.1 KQ434787 KZC05298.1 APGK01040737 APGK01040738 KB740984 KB631904 ENN76260.1 ERL87087.1 GFDF01003785 JAV10299.1 GFDF01003806 JAV10278.1 GDKW01000960 JAI55635.1 ACPB03001109 GAHY01000691 JAA76819.1 IAAA01001821 IAAA01001822 LAA03111.1 GL765107 EFZ16912.1 GFTR01003603 JAW12823.1 AAZX01000032 AAZX01016030 LRGB01001745 KZS10691.1 KB204017 ESO82123.1 DS231890 EDS43799.1 KQ759768 OAD62949.1 KK854094 PTY11165.1 GL732911 EFX63965.1 GFDL01008110 JAV26935.1 GDIP01094815 JAM08900.1 GU289394 ADK66883.1 GAPW01004256 JAC09342.1 CH477997 EAT34504.1 GG666677 EEN44211.1 HACG01038531 CEK85396.1 EAT34505.2 JXUM01131546 GAPW01003845 KQ567724 JAC09753.1 KXJ69305.1 GFDL01008252 JAV26793.1 GEFM01005894 JAP69902.1 JH817502 EKC28779.1 AMQN01004337 KB293237 ELU16260.1 GDIQ01019938 JAN74799.1 GGLE01001486 MBY05612.1 GFWV01006348 MAA31078.1 EAAA01002795 FR905707 CDQ81111.1 NEDP02005521 OWF39565.1 JH880525 ELR60903.1 KE164180 EPQ15773.1 KL517270 KFO88424.1 KL896018 KGL83122.1 AKHW03006215 KYO23451.1 AHAT01024887 KL217813 KFO99130.1 KL448313 KFO82555.1 KL409973 KFQ93042.1 MUZQ01000043 OWK61379.1 KL414671 KFW77973.1 KK735751 KFR15638.1 AGCU01069984 AGCU01069985 KL486402 KFO09741.1 HAEC01000112 SBQ68189.1 HAEF01006930 SBR44312.1 GBSH01002177 JAG66849.1 KL245982 KFV14843.1 HAEA01013013 SBQ41493.1 KK500860 KFP11139.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000027135
UP000215335
+ More
UP000192223 UP000053825 UP000242457 UP000053105 UP000005203 UP000007755 UP000036403 UP000005205 UP000075809 UP000053097 UP000279307 UP000078542 UP000078492 UP000076502 UP000019118 UP000030742 UP000015103 UP000002358 UP000076858 UP000030746 UP000002320 UP000007875 UP000085678 UP000000305 UP000008820 UP000001554 UP000245300 UP000076420 UP000069940 UP000249989 UP000005408 UP000014760 UP000087266 UP000008144 UP000265140 UP000193380 UP000261540 UP000192220 UP000242188 UP000261680 UP000002281 UP000001645 UP000261360 UP000261480 UP000261400 UP000053641 UP000050525 UP000018468 UP000054308 UP000053760 UP000053283 UP000197619 UP000002852 UP000053605 UP000007267 UP000261420 UP000261560 UP000261500 UP000001646 UP000053119
UP000192223 UP000053825 UP000242457 UP000053105 UP000005203 UP000007755 UP000036403 UP000005205 UP000075809 UP000053097 UP000279307 UP000078542 UP000078492 UP000076502 UP000019118 UP000030742 UP000015103 UP000002358 UP000076858 UP000030746 UP000002320 UP000007875 UP000085678 UP000000305 UP000008820 UP000001554 UP000245300 UP000076420 UP000069940 UP000249989 UP000005408 UP000014760 UP000087266 UP000008144 UP000265140 UP000193380 UP000261540 UP000192220 UP000242188 UP000261680 UP000002281 UP000001645 UP000261360 UP000261480 UP000261400 UP000053641 UP000050525 UP000018468 UP000054308 UP000053760 UP000053283 UP000197619 UP000002852 UP000053605 UP000007267 UP000261420 UP000261560 UP000261500 UP000001646 UP000053119
Interpro
CDD
ProteinModelPortal
A0A2W1BUQ7
A0A2H1WYQ7
A0A2A4K2R8
A0A194PRU1
A0A194RC43
A0A212ETT0
+ More
S4P930 A0A067R9C4 A0A1Y1KYF7 A0A232ENP5 A0A0B4UC52 A0A1W4XFX6 A0A0L7RA24 A0A2A3EL95 V9IIX3 A0A0N0BH50 A0A088A3U8 F4W7C1 A0A0J7KXD2 A0A158NPW7 A0A151X8V7 A0A026WIL4 A0A195CJ97 A0A195DSS0 A0A0K8SLD5 A0A154P250 N6U3U2 A0A1L8DV52 A0A1L8DV18 A0A0P4VRG0 R4G4K6 A0A2L2Y4L3 E9IRE0 A0A224XXD2 K7IXC5 A0A164TRN1 V3YVU6 B0WCS3 A0A310SXR9 H2Y4P3 A0A2R7VZ04 A0A1S3HQH2 E9HWB8 A0A1Q3FHB9 A0A0N8CVH9 E7BTP8 A0A023EJU0 Q16JQ5 C3ZTG4 S4RAH7 A0A2C9JWN1 A0A0B7AZA8 Q16JQ4 A0A023EKN9 A0A1Q3FH10 A0A131XU96 K1QC78 R7VJW8 A0A0P6HF76 A0A1S3PRE2 A0A2R5L800 A0A293LTH8 F6VMY2 A0A3P8XXR9 A0A060XNR3 A0A3B3RXK7 A0A2I4CVT3 A0A210PSV6 A0A384CRD0 L8IY39 F7B9P4 S7PXR1 G1N6Z1 A0A3B4XAX4 A0A3B3YHN5 A0A091GZ27 A0A3B5BC83 A0A099ZMY2 A0A151MG19 W5ME44 A0A091IHD3 A0A091GJI0 A0A091URK5 A0A218V634 A0A093PMB1 M3ZSX6 A0A091WL45 K7F802 A0A087VAU9 A0A3B4TIX6 A0A3B3BBX7 A0A1A8GCU5 A0A3B3TS66 H9GDR1 A0A3P8YA38 A0A1A8LI77 A0A0B8RPH2 A0A093CK25 A0A1A8E6A0 A0A091IR10
S4P930 A0A067R9C4 A0A1Y1KYF7 A0A232ENP5 A0A0B4UC52 A0A1W4XFX6 A0A0L7RA24 A0A2A3EL95 V9IIX3 A0A0N0BH50 A0A088A3U8 F4W7C1 A0A0J7KXD2 A0A158NPW7 A0A151X8V7 A0A026WIL4 A0A195CJ97 A0A195DSS0 A0A0K8SLD5 A0A154P250 N6U3U2 A0A1L8DV52 A0A1L8DV18 A0A0P4VRG0 R4G4K6 A0A2L2Y4L3 E9IRE0 A0A224XXD2 K7IXC5 A0A164TRN1 V3YVU6 B0WCS3 A0A310SXR9 H2Y4P3 A0A2R7VZ04 A0A1S3HQH2 E9HWB8 A0A1Q3FHB9 A0A0N8CVH9 E7BTP8 A0A023EJU0 Q16JQ5 C3ZTG4 S4RAH7 A0A2C9JWN1 A0A0B7AZA8 Q16JQ4 A0A023EKN9 A0A1Q3FH10 A0A131XU96 K1QC78 R7VJW8 A0A0P6HF76 A0A1S3PRE2 A0A2R5L800 A0A293LTH8 F6VMY2 A0A3P8XXR9 A0A060XNR3 A0A3B3RXK7 A0A2I4CVT3 A0A210PSV6 A0A384CRD0 L8IY39 F7B9P4 S7PXR1 G1N6Z1 A0A3B4XAX4 A0A3B3YHN5 A0A091GZ27 A0A3B5BC83 A0A099ZMY2 A0A151MG19 W5ME44 A0A091IHD3 A0A091GJI0 A0A091URK5 A0A218V634 A0A093PMB1 M3ZSX6 A0A091WL45 K7F802 A0A087VAU9 A0A3B4TIX6 A0A3B3BBX7 A0A1A8GCU5 A0A3B3TS66 H9GDR1 A0A3P8YA38 A0A1A8LI77 A0A0B8RPH2 A0A093CK25 A0A1A8E6A0 A0A091IR10
PDB
4DRZ
E-value=1.7751e-93,
Score=871
Ontologies
GO
GO:0005802
GO:0055037
GO:0005525
GO:0006895
GO:0045335
GO:0005829
GO:0003924
GO:0090382
GO:0042742
GO:0032456
GO:0004540
GO:0008033
GO:0090387
GO:0006886
GO:0005769
GO:0032482
GO:0032880
GO:0019003
GO:0008543
GO:0045995
GO:0031489
GO:0007264
GO:0005524
GO:0006419
GO:0000166
GO:0005737
GO:0016876
GO:0043039
GO:0004651
GO:0005634
PANTHER
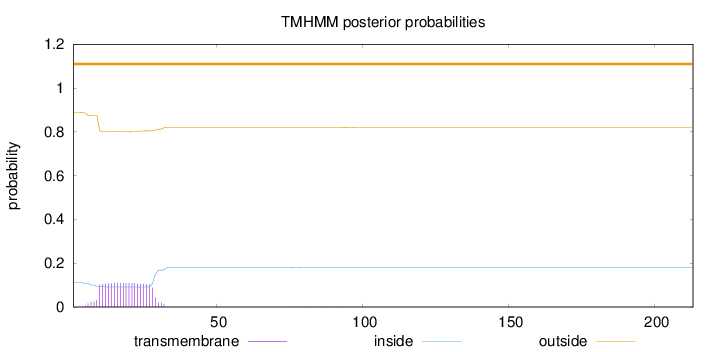
Topology
Length:
213
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.23623000000001
Exp number, first 60 AAs:
2.21964
Total prob of N-in:
0.11093
outside
1 - 213
Population Genetic Test Statistics
Pi
239.333748
Theta
177.472591
Tajima's D
1.212566
CLR
0
CSRT
0.72196390180491
Interpretation
Uncertain
