Gene
KWMTBOMO01933 Validated by peptides from experiments
Annotation
hypothetical_protein_KGM_08437_[Danaus_plexippus]
Full name
NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial
Location in the cell
Nuclear Reliability : 3.581
Sequence
CDS
ATGCTAAGAAGTACACCAGTATTATGTCGAACTTTGACGAACAAGATTGAAGATATTTCTACTCATACAGGCCAGAAATGGGATTCTGATGACTACAGGCTGGTAAGATTTACTAATGCGCCAAAACAAGTAAATCCGAATTGGGCCGTAAATTTAATTGCGGAAATACCTCCTAAAGAAGTAACTGAAAGAGTTGTTTGGTGTGATGGAGGTAGTGGCCCAGAGGGTCATCCACGAGTGTATATTAATTTAGATAAACCAGGTGACCATTCATGTGGATACTGTGGATTAAGATTCATAAAAAAAACTGGTCATTAA
Protein
MLRSTPVLCRTLTNKIEDISTHTGQKWDSDDYRLVRFTNAPKQVNPNWAVNLIAEIPPKEVTERVVWCDGGSGPEGHPRVYINLDKPGDHSCGYCGLRFIKKTGH
Summary
Description
Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone.
Similarity
Belongs to the complex I NDUFS6 subunit family.
Uniprot
A0A2W1BZ07
A0A2A4K2Q7
A0A2H1WYS3
A0A212ETU0
A0A194RD20
A0A194PLI0
+ More
S4P4L7 A0A2S2P5L8 A0A2P8YMS5 V5I811 A0A1S3D866 L7M645 A0A1I8NBD1 B3MLV5 C9W1L8 A0A131XK46 A0A224Z3X9 J3JXJ2 A0A131YTS6 A0A0N8APX4 D6WLU6 N6U2V5 A0A0P4VPK1 R4FK98 A0A1D2NC77 A0A067QI11 B4KL31 A0A0P6CUG9 A0A1B0FH78 A0A0P5LMS4 A0A0K8TSS9 A0A0P6ANZ3 A0A2H8TIV7 A0A1A9Z7W4 A0A1L8EIB3 A0A1I8Q5X0 A0A1W4UHL1 A0A2R5LDT2 A0A1Y1MQE5 A0A1B6L7X8 A0A1A9V890 A0A026WKZ6 A0A1A9WKK8 A0A1L8E0U7 J9JKP8 A0A3L8DBY4 E9GNF2 A0A023FN28 A0A023FR16 A0A1A9XEQ3 A0A1E1WYL3 C4WS83 A0A0C9SAG0 A0A293MDZ0 B4N0D8 B3N4T0 A0A0L0CMD6 A0A2S2QJ69 A0A1B0D2I7 A0A0J7KIT8 A0A0J9QW16 A0A1S3IE50 A0A0P5FQ78 B4Q3I9 Q9VMU0 A0A0A1WRS8 B4I1D4 A0A1B6C7M3 E0VEN9 A0A3B0JIN8 B7P5E4 A0A1B6FII1 B4P3S8 W8BWA1 B4G8M4 F4WNJ9 B4JDB4 A0A034WMY6 A0A0C9RUG6 A0A2A3E0J7 V9IKP8 A0A1B6FLE8 A0A0M4E8V3 A0A1B6ENW0 A0A151WVN8 A0A2R7W5K1 Q29MW6 A0A0P4ZZW9 E2A1K8 A0A0K8U1Y4 A0A088AHE2 E2B866 A0A154PFL2 A0A131Y1Z3 A0A0L7QXP7 A0A0K8RBZ3 A0A195FW77 B4LUR4 E9J097
S4P4L7 A0A2S2P5L8 A0A2P8YMS5 V5I811 A0A1S3D866 L7M645 A0A1I8NBD1 B3MLV5 C9W1L8 A0A131XK46 A0A224Z3X9 J3JXJ2 A0A131YTS6 A0A0N8APX4 D6WLU6 N6U2V5 A0A0P4VPK1 R4FK98 A0A1D2NC77 A0A067QI11 B4KL31 A0A0P6CUG9 A0A1B0FH78 A0A0P5LMS4 A0A0K8TSS9 A0A0P6ANZ3 A0A2H8TIV7 A0A1A9Z7W4 A0A1L8EIB3 A0A1I8Q5X0 A0A1W4UHL1 A0A2R5LDT2 A0A1Y1MQE5 A0A1B6L7X8 A0A1A9V890 A0A026WKZ6 A0A1A9WKK8 A0A1L8E0U7 J9JKP8 A0A3L8DBY4 E9GNF2 A0A023FN28 A0A023FR16 A0A1A9XEQ3 A0A1E1WYL3 C4WS83 A0A0C9SAG0 A0A293MDZ0 B4N0D8 B3N4T0 A0A0L0CMD6 A0A2S2QJ69 A0A1B0D2I7 A0A0J7KIT8 A0A0J9QW16 A0A1S3IE50 A0A0P5FQ78 B4Q3I9 Q9VMU0 A0A0A1WRS8 B4I1D4 A0A1B6C7M3 E0VEN9 A0A3B0JIN8 B7P5E4 A0A1B6FII1 B4P3S8 W8BWA1 B4G8M4 F4WNJ9 B4JDB4 A0A034WMY6 A0A0C9RUG6 A0A2A3E0J7 V9IKP8 A0A1B6FLE8 A0A0M4E8V3 A0A1B6ENW0 A0A151WVN8 A0A2R7W5K1 Q29MW6 A0A0P4ZZW9 E2A1K8 A0A0K8U1Y4 A0A088AHE2 E2B866 A0A154PFL2 A0A131Y1Z3 A0A0L7QXP7 A0A0K8RBZ3 A0A195FW77 B4LUR4 E9J097
Pubmed
28756777
22118469
26354079
23622113
29403074
25576852
+ More
25315136 17994087 20650005 24029695 28049606 28797301 22516182 26830274 18362917 19820115 23537049 27129103 27289101 24845553 26369729 28004739 24508170 30249741 21292972 28503490 26131772 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 20566863 17550304 24495485 21719571 25348373 15632085 20798317 21282665
25315136 17994087 20650005 24029695 28049606 28797301 22516182 26830274 18362917 19820115 23537049 27129103 27289101 24845553 26369729 28004739 24508170 30249741 21292972 28503490 26131772 26108605 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 20566863 17550304 24495485 21719571 25348373 15632085 20798317 21282665
EMBL
KZ149895
PZC78814.1
NWSH01000193
PCG78541.1
ODYU01012086
SOQ58201.1
+ More
AGBW02012530 OWR44910.1 KQ460398 KPJ15170.1 KQ459601 KPI93853.1 GAIX01005914 JAA86646.1 GGMR01012148 MBY24767.1 PYGN01000485 PSN45546.1 GALX01005495 JAB62971.1 GACK01006421 JAA58613.1 CH902620 EDV31783.1 EZ406169 ACX53965.1 GEFH01001644 JAP66937.1 GFPF01010117 MAA21263.1 BT127961 AEE62923.1 GEDV01006605 JAP81952.1 GDIQ01254467 LRGB01002190 JAJ97257.1 KZS08532.1 KQ971343 EFA04681.1 APGK01051866 KB741207 KB632327 ENN72902.1 ERL92519.1 GDKW01002168 JAI54427.1 ACPB03005117 GAHY01001953 JAA75557.1 LJIJ01000094 ODN02839.1 KK853366 KDR08107.1 CH933807 EDW12781.1 GDIQ01088370 JAN06367.1 CCAG010002037 GDIQ01192126 JAK59599.1 GDAI01000174 JAI17429.1 GDIP01026776 JAM76939.1 GFXV01002268 MBW14073.1 GFDG01000378 JAV18421.1 GGLE01003371 MBY07497.1 GEZM01024260 JAV87821.1 GEBQ01020242 JAT19735.1 KK107167 EZA56346.1 GFDF01001721 JAV12363.1 ABLF02024711 QOIP01000010 RLU17418.1 GL732554 EFX79018.1 GBBK01001326 JAC23156.1 GBBK01001324 JAC23158.1 GFAC01007093 JAT92095.1 AK340083 BAH70753.1 GBZX01002613 JAG90127.1 GFWV01014454 MAA39183.1 CH963920 EDW77551.1 CH954177 EDV57832.1 JRES01000186 KNC33513.1 GGMS01008540 MBY77743.1 AJVK01022836 LBMM01007019 KMQ90116.1 CM002910 KMY88257.1 GDIQ01259440 JAJ92284.1 CM000361 EDX03793.1 AE014134 BT011535 AAF52221.1 AAS15671.1 GBXI01012523 JAD01769.1 CH480820 EDW54341.1 GEDC01027816 JAS09482.1 DS235093 EEB11845.1 OUUW01000006 SPP82237.1 ABJB010209339 DS640379 EEC01816.1 GECZ01019815 JAS49954.1 CM000157 EDW88379.1 GAMC01000955 JAC05601.1 CH479180 EDW28704.1 GL888237 EGI64192.1 CH916368 EDW03287.1 GAKP01003417 JAC55535.1 GBYB01012485 JAG82252.1 KZ288478 PBC25275.1 JR051996 AEY61605.1 GECZ01018770 JAS50999.1 CP012523 ALC38501.1 GECZ01030149 JAS39620.1 KQ982706 KYQ51876.1 KK854307 PTY14441.1 CH379060 EAL33577.2 GDIP01205804 JAJ17598.1 GL435766 EFN72681.1 GDHF01031979 GDHF01023074 GDHF01019564 GDHF01015786 JAI20335.1 JAI29240.1 JAI32750.1 JAI36528.1 GL446286 EFN88068.1 KQ434893 KZC10602.1 GEFM01003278 JAP72518.1 KQ414700 KOC63385.1 GADI01005425 JAA68383.1 KQ981215 KYN44572.1 CH940649 EDW64241.1 GL767355 EFZ13756.1
AGBW02012530 OWR44910.1 KQ460398 KPJ15170.1 KQ459601 KPI93853.1 GAIX01005914 JAA86646.1 GGMR01012148 MBY24767.1 PYGN01000485 PSN45546.1 GALX01005495 JAB62971.1 GACK01006421 JAA58613.1 CH902620 EDV31783.1 EZ406169 ACX53965.1 GEFH01001644 JAP66937.1 GFPF01010117 MAA21263.1 BT127961 AEE62923.1 GEDV01006605 JAP81952.1 GDIQ01254467 LRGB01002190 JAJ97257.1 KZS08532.1 KQ971343 EFA04681.1 APGK01051866 KB741207 KB632327 ENN72902.1 ERL92519.1 GDKW01002168 JAI54427.1 ACPB03005117 GAHY01001953 JAA75557.1 LJIJ01000094 ODN02839.1 KK853366 KDR08107.1 CH933807 EDW12781.1 GDIQ01088370 JAN06367.1 CCAG010002037 GDIQ01192126 JAK59599.1 GDAI01000174 JAI17429.1 GDIP01026776 JAM76939.1 GFXV01002268 MBW14073.1 GFDG01000378 JAV18421.1 GGLE01003371 MBY07497.1 GEZM01024260 JAV87821.1 GEBQ01020242 JAT19735.1 KK107167 EZA56346.1 GFDF01001721 JAV12363.1 ABLF02024711 QOIP01000010 RLU17418.1 GL732554 EFX79018.1 GBBK01001326 JAC23156.1 GBBK01001324 JAC23158.1 GFAC01007093 JAT92095.1 AK340083 BAH70753.1 GBZX01002613 JAG90127.1 GFWV01014454 MAA39183.1 CH963920 EDW77551.1 CH954177 EDV57832.1 JRES01000186 KNC33513.1 GGMS01008540 MBY77743.1 AJVK01022836 LBMM01007019 KMQ90116.1 CM002910 KMY88257.1 GDIQ01259440 JAJ92284.1 CM000361 EDX03793.1 AE014134 BT011535 AAF52221.1 AAS15671.1 GBXI01012523 JAD01769.1 CH480820 EDW54341.1 GEDC01027816 JAS09482.1 DS235093 EEB11845.1 OUUW01000006 SPP82237.1 ABJB010209339 DS640379 EEC01816.1 GECZ01019815 JAS49954.1 CM000157 EDW88379.1 GAMC01000955 JAC05601.1 CH479180 EDW28704.1 GL888237 EGI64192.1 CH916368 EDW03287.1 GAKP01003417 JAC55535.1 GBYB01012485 JAG82252.1 KZ288478 PBC25275.1 JR051996 AEY61605.1 GECZ01018770 JAS50999.1 CP012523 ALC38501.1 GECZ01030149 JAS39620.1 KQ982706 KYQ51876.1 KK854307 PTY14441.1 CH379060 EAL33577.2 GDIP01205804 JAJ17598.1 GL435766 EFN72681.1 GDHF01031979 GDHF01023074 GDHF01019564 GDHF01015786 JAI20335.1 JAI29240.1 JAI32750.1 JAI36528.1 GL446286 EFN88068.1 KQ434893 KZC10602.1 GEFM01003278 JAP72518.1 KQ414700 KOC63385.1 GADI01005425 JAA68383.1 KQ981215 KYN44572.1 CH940649 EDW64241.1 GL767355 EFZ13756.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000245037
UP000079169
+ More
UP000095301 UP000007801 UP000076858 UP000007266 UP000019118 UP000030742 UP000015103 UP000094527 UP000027135 UP000009192 UP000092444 UP000092445 UP000095300 UP000192221 UP000078200 UP000053097 UP000091820 UP000007819 UP000279307 UP000000305 UP000092443 UP000007798 UP000008711 UP000037069 UP000092462 UP000036403 UP000085678 UP000000304 UP000000803 UP000001292 UP000009046 UP000268350 UP000001555 UP000002282 UP000008744 UP000007755 UP000001070 UP000242457 UP000092553 UP000075809 UP000001819 UP000000311 UP000005203 UP000008237 UP000076502 UP000053825 UP000078541 UP000008792
UP000095301 UP000007801 UP000076858 UP000007266 UP000019118 UP000030742 UP000015103 UP000094527 UP000027135 UP000009192 UP000092444 UP000092445 UP000095300 UP000192221 UP000078200 UP000053097 UP000091820 UP000007819 UP000279307 UP000000305 UP000092443 UP000007798 UP000008711 UP000037069 UP000092462 UP000036403 UP000085678 UP000000304 UP000000803 UP000001292 UP000009046 UP000268350 UP000001555 UP000002282 UP000008744 UP000007755 UP000001070 UP000242457 UP000092553 UP000075809 UP000001819 UP000000311 UP000005203 UP000008237 UP000076502 UP000053825 UP000078541 UP000008792
PRIDE
Pfam
PF10276 zf-CHCC
ProteinModelPortal
A0A2W1BZ07
A0A2A4K2Q7
A0A2H1WYS3
A0A212ETU0
A0A194RD20
A0A194PLI0
+ More
S4P4L7 A0A2S2P5L8 A0A2P8YMS5 V5I811 A0A1S3D866 L7M645 A0A1I8NBD1 B3MLV5 C9W1L8 A0A131XK46 A0A224Z3X9 J3JXJ2 A0A131YTS6 A0A0N8APX4 D6WLU6 N6U2V5 A0A0P4VPK1 R4FK98 A0A1D2NC77 A0A067QI11 B4KL31 A0A0P6CUG9 A0A1B0FH78 A0A0P5LMS4 A0A0K8TSS9 A0A0P6ANZ3 A0A2H8TIV7 A0A1A9Z7W4 A0A1L8EIB3 A0A1I8Q5X0 A0A1W4UHL1 A0A2R5LDT2 A0A1Y1MQE5 A0A1B6L7X8 A0A1A9V890 A0A026WKZ6 A0A1A9WKK8 A0A1L8E0U7 J9JKP8 A0A3L8DBY4 E9GNF2 A0A023FN28 A0A023FR16 A0A1A9XEQ3 A0A1E1WYL3 C4WS83 A0A0C9SAG0 A0A293MDZ0 B4N0D8 B3N4T0 A0A0L0CMD6 A0A2S2QJ69 A0A1B0D2I7 A0A0J7KIT8 A0A0J9QW16 A0A1S3IE50 A0A0P5FQ78 B4Q3I9 Q9VMU0 A0A0A1WRS8 B4I1D4 A0A1B6C7M3 E0VEN9 A0A3B0JIN8 B7P5E4 A0A1B6FII1 B4P3S8 W8BWA1 B4G8M4 F4WNJ9 B4JDB4 A0A034WMY6 A0A0C9RUG6 A0A2A3E0J7 V9IKP8 A0A1B6FLE8 A0A0M4E8V3 A0A1B6ENW0 A0A151WVN8 A0A2R7W5K1 Q29MW6 A0A0P4ZZW9 E2A1K8 A0A0K8U1Y4 A0A088AHE2 E2B866 A0A154PFL2 A0A131Y1Z3 A0A0L7QXP7 A0A0K8RBZ3 A0A195FW77 B4LUR4 E9J097
S4P4L7 A0A2S2P5L8 A0A2P8YMS5 V5I811 A0A1S3D866 L7M645 A0A1I8NBD1 B3MLV5 C9W1L8 A0A131XK46 A0A224Z3X9 J3JXJ2 A0A131YTS6 A0A0N8APX4 D6WLU6 N6U2V5 A0A0P4VPK1 R4FK98 A0A1D2NC77 A0A067QI11 B4KL31 A0A0P6CUG9 A0A1B0FH78 A0A0P5LMS4 A0A0K8TSS9 A0A0P6ANZ3 A0A2H8TIV7 A0A1A9Z7W4 A0A1L8EIB3 A0A1I8Q5X0 A0A1W4UHL1 A0A2R5LDT2 A0A1Y1MQE5 A0A1B6L7X8 A0A1A9V890 A0A026WKZ6 A0A1A9WKK8 A0A1L8E0U7 J9JKP8 A0A3L8DBY4 E9GNF2 A0A023FN28 A0A023FR16 A0A1A9XEQ3 A0A1E1WYL3 C4WS83 A0A0C9SAG0 A0A293MDZ0 B4N0D8 B3N4T0 A0A0L0CMD6 A0A2S2QJ69 A0A1B0D2I7 A0A0J7KIT8 A0A0J9QW16 A0A1S3IE50 A0A0P5FQ78 B4Q3I9 Q9VMU0 A0A0A1WRS8 B4I1D4 A0A1B6C7M3 E0VEN9 A0A3B0JIN8 B7P5E4 A0A1B6FII1 B4P3S8 W8BWA1 B4G8M4 F4WNJ9 B4JDB4 A0A034WMY6 A0A0C9RUG6 A0A2A3E0J7 V9IKP8 A0A1B6FLE8 A0A0M4E8V3 A0A1B6ENW0 A0A151WVN8 A0A2R7W5K1 Q29MW6 A0A0P4ZZW9 E2A1K8 A0A0K8U1Y4 A0A088AHE2 E2B866 A0A154PFL2 A0A131Y1Z3 A0A0L7QXP7 A0A0K8RBZ3 A0A195FW77 B4LUR4 E9J097
PDB
5GUP
E-value=3.46174e-23,
Score=261
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion inner membrane
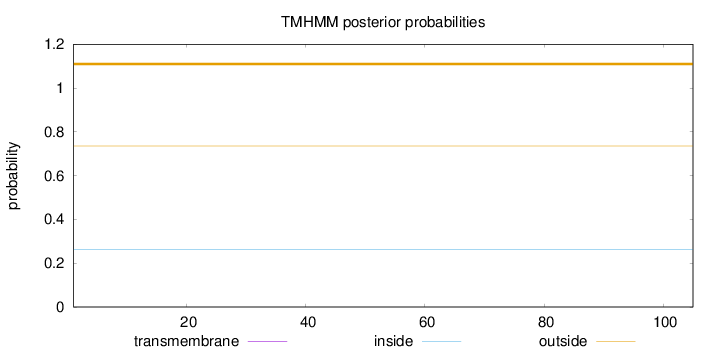
Length:
105
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.26428
outside
1 - 105
Population Genetic Test Statistics
Pi
177.941181
Theta
159.845167
Tajima's D
0.346191
CLR
1.101758
CSRT
0.467526623668817
Interpretation
Uncertain
