Gene
KWMTBOMO01930
Pre Gene Modal
BGIBMGA006218
Annotation
PREDICTED:_kinesin-associated_protein_3_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.112
Sequence
CDS
ATGAGTCCCGTGGAAAGTGGTCCTGTTGCTGTTTTGCACCATGTTGATGATTATGTAGAATTATTATATGATGATATACCTGAAAAAATTAAAGGATCAGCACTCATTCTACAACTTGCTAGAAACCCTGATAATCTTTTAGAGTTGGCTAGAAATGAAGCCTTACTTAGTGCCCTGTCCAGAGTCCTGAGAGAAGAATGGAAACGAAGCATTGAACTAAGTACCAATATTGTATACACATTTTTTTGTTTCTCAACCTACAATGAATTTCATCCAGTTATTATACAATACAAAATAGGATCATTGTGTATGGATGTAATAGATTATGAATTAAAAAGATATGATCAATGGAAAGAAAAAGTTGAAGGAAAAAAATCTATACTAACCCCTGATAAAAGCGAACTCCCAAAAAGTAGGATTCCCGAACCAAAGCGACGACCTAAATCAGGCACTTGGGCTGTTGCAGATGTGAATTTGCAGAAATCTAGATCTCTAGTATCTAGCTATCATGAAGAGCTGTGTAGTGCTTCAGATGAGTGCCTTTCTAAAAGTGATGAAGAACAATTGAAAAGAAAGCTTAAAACTTTGTCAAAGCGACAAGAGCAACTTTTAAGAGTTGCATTTTACATGTTACTCAATATTGCAGATAATGTAAAGGTTGAAGAGAAGATGCATAAAAAGGAAATAGTTGGTCTTTTAATTGGTGCCATGGAAAGGCATTCAAATATTGATTTACTTATACTTATTGTGTCCTTCCTTCAAAAGCTTTCAATTTTTGTCGAAAACAAAAATAGCATGGCTTCTCGAGCCATTATAGAAAAGTTAGCACCCTTATTGGATTCATCAAATGCTGATCTTGTAAATGTAACTTTAAAGTTATTATTTAACCTTACTTTTGACACAAAATTACGCAATAAAATGATTAAACTTGGACTATTACCAAAGTTCGTCCAATTTACTAGTGATGAAAAACATATAAATTTAGCAATGAAAATACTCTACCATTTGAGTATGGATGATCGCGTTAAGATAATGTTTACACAGTCAGACTGTGTAAAATTGTTAACGGACATGCTGCTGTTGAATGTCAATGCAGAATCGGGAGGCTCTACAGATGTTTTGTTGGCATTATGTGTAAACCTTGCTTGGTGTGAGCGAGCAGCGCACCAAATGGCTGCTGAAGGCCGACTTCGTGAACTATTGGCACGTGCTTTCCGGTATTGCAATTCTGTACTCATGAAACTTGTTCGGAACCTTTCCCATCATCCACAAAACAAACCACTTTTTTTGGAATTCGTCGGAGATATCGCGGGAGCCGTAACTAGTGGCGAAACTAGTGAGGAGTTTCACATTGAATGCATGGGGACTCTCAGCAACATACTCAATCCAGAAAATATAGATATATATGCTGTAGTAGAAAGATATAATCTGATACCTTGCATCATGAAAATTTTGGATCCAGAGAGCGGGAGCAGTGAAGAGTTATTACTGGAGGGCGTGGTGATGGCGGGCGCAGTGGCGGCTGAGTCGCGGGCGGCGGGCGCGCTGGTGCGTGCGGGCGCGGGGCCGGCGCTCGTGGTCGCGCTGCGCACGCATCAGGCCGACGACGAACACGTGTTGCAAACTGTCTTCGCCTTTCGACAGCTGCTCTCGCATCCCAAAGCGGCCGAACACCTCGTCTCGCAGACAGAAGCTCCTGCATATCTCATTGATCTGATGCAAGATAAGAATGCTGAAATCAGGAAAATGTGTGACTCTTGCCTCGATATTATATCACAAATGCAGAACGAATGGGCGGCCCGAATAAAGGTATGCATGGTATACATAAATTAA
Protein
MSPVESGPVAVLHHVDDYVELLYDDIPEKIKGSALILQLARNPDNLLELARNEALLSALSRVLREEWKRSIELSTNIVYTFFCFSTYNEFHPVIIQYKIGSLCMDVIDYELKRYDQWKEKVEGKKSILTPDKSELPKSRIPEPKRRPKSGTWAVADVNLQKSRSLVSSYHEELCSASDECLSKSDEEQLKRKLKTLSKRQEQLLRVAFYMLLNIADNVKVEEKMHKKEIVGLLIGAMERHSNIDLLILIVSFLQKLSIFVENKNSMASRAIIEKLAPLLDSSNADLVNVTLKLLFNLTFDTKLRNKMIKLGLLPKFVQFTSDEKHINLAMKILYHLSMDDRVKIMFTQSDCVKLLTDMLLLNVNAESGGSTDVLLALCVNLAWCERAAHQMAAEGRLRELLARAFRYCNSVLMKLVRNLSHHPQNKPLFLEFVGDIAGAVTSGETSEEFHIECMGTLSNILNPENIDIYAVVERYNLIPCIMKILDPESGSSEELLLEGVVMAGAVAAESRAAGALVRAGAGPALVVALRTHQADDEHVLQTVFAFRQLLSHPKAAEHLVSQTEAPAYLIDLMQDKNAEIRKMCDSCLDIISQMQNEWAARIKVCMVYIN
Summary
Uniprot
H9J9M5
A0A2A4K3Q0
A0A2W1C1G7
A0A194PKK2
A0A0L7LN66
A0A194RCH7
+ More
A0A212ETY8 A0A2H1WIV4 A0A023EXS2 A0A1B0D2V3 Q17J47 A0A1B0C902 A0A1Q3FAC7 A0A1Q3FAC1 A0A1L8DUQ5 A0A1Q3FAA0 A0A1L8DUQ1 B0WM84 A0A1L8DVG0 A0A1Q3FA95 A0A1L8DVH1 A0A1A9VIK3 A0A182G522 A0A1B0BHR2 A0A1B0A9Z5 A0A1A9WCC5 A0A1B0FQ22 A0A1I8MFF7 A0A1A9XC32 A0A0A1X335 W8BVT7 A0A0K8UF20 A0A1I8MFD8 A0A1I8MFE0 W8AWE6 W8BJ22 A0A034VN10 A0A1J1IPX6 A0A182IQU9 A0A0A1XEH2 A0A0A1WLD8 A0A0K8WLV1 A0A0K8TVC2 A0A084WPG0 A0A0L0BNH1 A0A1I8Q823 A0A182M7G2 A0A182R7I5 B4NES2 A0A336KQW8 A0A1I8Q864 A0A182QJ16 A0A182YQ66 A0A182V6E4 A0A182K7A1 A0A1I8Q867 A0A1I8Q836 Q7QEW8 A0A182X7Z9 A0A182HIB8 A0A182N1L0 B4PYC9 A0A0P8XL03 B3MVZ7 B3NV80 A0A1W4V242 B7Z139 A8JUR4 B4R2S8 A0A3B0J785 A0A182P2A7 M9PGW1 Q9VZ07 A0A182W502 B5DM22 A0A182HBS8 A8JUR5 A0A0M4EXV3 A0A3B0JC21 B4JL35 A0A0K8W6T2 D6WK33 B4L7I2 A0A2P8ZFZ3 A0A2M4CS53 A0A0Q9VZE0 A0A2M4CTK2 A0A232FJW3 A0A2M4CSA3 A0A0Q9VZF3 A0A2M4A7P2 A0A2M4A7L9 K7IWB7 E2APE2 E9IPB0 F4X2C7 A0A0C9PRJ8 E2BKQ4 A0A151XGA3
A0A212ETY8 A0A2H1WIV4 A0A023EXS2 A0A1B0D2V3 Q17J47 A0A1B0C902 A0A1Q3FAC7 A0A1Q3FAC1 A0A1L8DUQ5 A0A1Q3FAA0 A0A1L8DUQ1 B0WM84 A0A1L8DVG0 A0A1Q3FA95 A0A1L8DVH1 A0A1A9VIK3 A0A182G522 A0A1B0BHR2 A0A1B0A9Z5 A0A1A9WCC5 A0A1B0FQ22 A0A1I8MFF7 A0A1A9XC32 A0A0A1X335 W8BVT7 A0A0K8UF20 A0A1I8MFD8 A0A1I8MFE0 W8AWE6 W8BJ22 A0A034VN10 A0A1J1IPX6 A0A182IQU9 A0A0A1XEH2 A0A0A1WLD8 A0A0K8WLV1 A0A0K8TVC2 A0A084WPG0 A0A0L0BNH1 A0A1I8Q823 A0A182M7G2 A0A182R7I5 B4NES2 A0A336KQW8 A0A1I8Q864 A0A182QJ16 A0A182YQ66 A0A182V6E4 A0A182K7A1 A0A1I8Q867 A0A1I8Q836 Q7QEW8 A0A182X7Z9 A0A182HIB8 A0A182N1L0 B4PYC9 A0A0P8XL03 B3MVZ7 B3NV80 A0A1W4V242 B7Z139 A8JUR4 B4R2S8 A0A3B0J785 A0A182P2A7 M9PGW1 Q9VZ07 A0A182W502 B5DM22 A0A182HBS8 A8JUR5 A0A0M4EXV3 A0A3B0JC21 B4JL35 A0A0K8W6T2 D6WK33 B4L7I2 A0A2P8ZFZ3 A0A2M4CS53 A0A0Q9VZE0 A0A2M4CTK2 A0A232FJW3 A0A2M4CSA3 A0A0Q9VZF3 A0A2M4A7P2 A0A2M4A7L9 K7IWB7 E2APE2 E9IPB0 F4X2C7 A0A0C9PRJ8 E2BKQ4 A0A151XGA3
Pubmed
19121390
28756777
26354079
26227816
22118469
24945155
+ More
17510324 26483478 25315136 25830018 24495485 25348373 24438588 26108605 17994087 25244985 12364791 14747013 17210077 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 29403074 28648823 20075255 20798317 21282665 21719571
17510324 26483478 25315136 25830018 24495485 25348373 24438588 26108605 17994087 25244985 12364791 14747013 17210077 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18362917 19820115 29403074 28648823 20075255 20798317 21282665 21719571
EMBL
BABH01005066
BABH01005067
NWSH01000193
PCG78538.1
KZ149895
PZC78816.1
+ More
KQ459601 KPI93857.1 JTDY01000501 KOB76892.1 KQ460398 KPJ15174.1 AGBW02012530 OWR44914.1 ODYU01008865 SOQ52832.1 GAPW01000289 JAC13309.1 AJVK01002869 AJVK01002870 CH477235 EAT46662.1 AJWK01001674 GFDL01010573 JAV24472.1 GFDL01010558 JAV24487.1 GFDF01003920 JAV10164.1 GFDL01010596 JAV24449.1 GFDF01003921 JAV10163.1 DS231995 EDS30918.1 GFDF01003672 JAV10412.1 GFDL01010541 JAV24504.1 GFDF01003671 JAV10413.1 JXUM01042847 JXUM01042848 KQ561341 KXJ78911.1 JXJN01014547 CCAG010019316 GBXI01009137 JAD05155.1 GAMC01013281 JAB93274.1 GDHF01027374 JAI24940.1 GAMC01013280 JAB93275.1 GAMC01013279 GAMC01013278 JAB93276.1 GAKP01016004 JAC42948.1 CVRI01000057 CRL02271.1 GBXI01004925 JAD09367.1 GBXI01014791 JAC99500.1 GDHF01000252 JAI52062.1 GDHF01033880 JAI18434.1 ATLV01025075 KE525369 KFB52104.1 JRES01001701 KNC20809.1 AXCM01001441 CH964239 EDW82241.2 UFQS01000842 UFQT01000842 SSX07216.1 SSX27559.1 AXCN02002448 AAAB01008846 EAA06454.5 APCN01003925 CM000162 EDX01991.1 CH902625 KPU75451.1 EDV35142.1 CH954180 EDV46068.1 AE014298 ACL82918.1 ABW09390.2 CM000366 EDX17627.1 OUUW01000003 SPP78084.1 AGB95288.2 BT010070 AAF48025.4 AAQ22539.1 CH379064 EDY72539.1 JXUM01125670 JXUM01125671 JXUM01125672 ABW09391.3 CP012528 ALC48680.1 SPP78083.1 CH916370 EDW00288.1 GDHF01005547 GDHF01002746 JAI46767.1 JAI49568.1 KQ971342 EFA03622.2 CH933813 EDW10976.2 PYGN01000069 PSN55423.1 GGFL01003982 MBW68160.1 CH940677 KRF78217.1 GGFL01003980 MBW68158.1 NNAY01000110 OXU30873.1 GGFL01003981 MBW68159.1 KRF78218.1 GGFK01003454 MBW36775.1 GGFK01003463 MBW36784.1 GL441524 EFN64687.1 GL764479 EFZ17577.1 GL888576 EGI59397.1 GBYB01003953 JAG73720.1 GL448819 EFN83752.1 KQ982179 KYQ59280.1
KQ459601 KPI93857.1 JTDY01000501 KOB76892.1 KQ460398 KPJ15174.1 AGBW02012530 OWR44914.1 ODYU01008865 SOQ52832.1 GAPW01000289 JAC13309.1 AJVK01002869 AJVK01002870 CH477235 EAT46662.1 AJWK01001674 GFDL01010573 JAV24472.1 GFDL01010558 JAV24487.1 GFDF01003920 JAV10164.1 GFDL01010596 JAV24449.1 GFDF01003921 JAV10163.1 DS231995 EDS30918.1 GFDF01003672 JAV10412.1 GFDL01010541 JAV24504.1 GFDF01003671 JAV10413.1 JXUM01042847 JXUM01042848 KQ561341 KXJ78911.1 JXJN01014547 CCAG010019316 GBXI01009137 JAD05155.1 GAMC01013281 JAB93274.1 GDHF01027374 JAI24940.1 GAMC01013280 JAB93275.1 GAMC01013279 GAMC01013278 JAB93276.1 GAKP01016004 JAC42948.1 CVRI01000057 CRL02271.1 GBXI01004925 JAD09367.1 GBXI01014791 JAC99500.1 GDHF01000252 JAI52062.1 GDHF01033880 JAI18434.1 ATLV01025075 KE525369 KFB52104.1 JRES01001701 KNC20809.1 AXCM01001441 CH964239 EDW82241.2 UFQS01000842 UFQT01000842 SSX07216.1 SSX27559.1 AXCN02002448 AAAB01008846 EAA06454.5 APCN01003925 CM000162 EDX01991.1 CH902625 KPU75451.1 EDV35142.1 CH954180 EDV46068.1 AE014298 ACL82918.1 ABW09390.2 CM000366 EDX17627.1 OUUW01000003 SPP78084.1 AGB95288.2 BT010070 AAF48025.4 AAQ22539.1 CH379064 EDY72539.1 JXUM01125670 JXUM01125671 JXUM01125672 ABW09391.3 CP012528 ALC48680.1 SPP78083.1 CH916370 EDW00288.1 GDHF01005547 GDHF01002746 JAI46767.1 JAI49568.1 KQ971342 EFA03622.2 CH933813 EDW10976.2 PYGN01000069 PSN55423.1 GGFL01003982 MBW68160.1 CH940677 KRF78217.1 GGFL01003980 MBW68158.1 NNAY01000110 OXU30873.1 GGFL01003981 MBW68159.1 KRF78218.1 GGFK01003454 MBW36775.1 GGFK01003463 MBW36784.1 GL441524 EFN64687.1 GL764479 EFZ17577.1 GL888576 EGI59397.1 GBYB01003953 JAG73720.1 GL448819 EFN83752.1 KQ982179 KYQ59280.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000037510
UP000053240
UP000007151
+ More
UP000092462 UP000008820 UP000092461 UP000002320 UP000078200 UP000069940 UP000249989 UP000092460 UP000092445 UP000091820 UP000092444 UP000095301 UP000092443 UP000183832 UP000075880 UP000030765 UP000037069 UP000095300 UP000075883 UP000075900 UP000007798 UP000075886 UP000076408 UP000075903 UP000075881 UP000007062 UP000076407 UP000075840 UP000075884 UP000002282 UP000007801 UP000008711 UP000192221 UP000000803 UP000000304 UP000268350 UP000075885 UP000075920 UP000001819 UP000092553 UP000001070 UP000007266 UP000009192 UP000245037 UP000008792 UP000215335 UP000002358 UP000000311 UP000007755 UP000008237 UP000075809
UP000092462 UP000008820 UP000092461 UP000002320 UP000078200 UP000069940 UP000249989 UP000092460 UP000092445 UP000091820 UP000092444 UP000095301 UP000092443 UP000183832 UP000075880 UP000030765 UP000037069 UP000095300 UP000075883 UP000075900 UP000007798 UP000075886 UP000076408 UP000075903 UP000075881 UP000007062 UP000076407 UP000075840 UP000075884 UP000002282 UP000007801 UP000008711 UP000192221 UP000000803 UP000000304 UP000268350 UP000075885 UP000075920 UP000001819 UP000092553 UP000001070 UP000007266 UP000009192 UP000245037 UP000008792 UP000215335 UP000002358 UP000000311 UP000007755 UP000008237 UP000075809
Interpro
Gene 3D
ProteinModelPortal
H9J9M5
A0A2A4K3Q0
A0A2W1C1G7
A0A194PKK2
A0A0L7LN66
A0A194RCH7
+ More
A0A212ETY8 A0A2H1WIV4 A0A023EXS2 A0A1B0D2V3 Q17J47 A0A1B0C902 A0A1Q3FAC7 A0A1Q3FAC1 A0A1L8DUQ5 A0A1Q3FAA0 A0A1L8DUQ1 B0WM84 A0A1L8DVG0 A0A1Q3FA95 A0A1L8DVH1 A0A1A9VIK3 A0A182G522 A0A1B0BHR2 A0A1B0A9Z5 A0A1A9WCC5 A0A1B0FQ22 A0A1I8MFF7 A0A1A9XC32 A0A0A1X335 W8BVT7 A0A0K8UF20 A0A1I8MFD8 A0A1I8MFE0 W8AWE6 W8BJ22 A0A034VN10 A0A1J1IPX6 A0A182IQU9 A0A0A1XEH2 A0A0A1WLD8 A0A0K8WLV1 A0A0K8TVC2 A0A084WPG0 A0A0L0BNH1 A0A1I8Q823 A0A182M7G2 A0A182R7I5 B4NES2 A0A336KQW8 A0A1I8Q864 A0A182QJ16 A0A182YQ66 A0A182V6E4 A0A182K7A1 A0A1I8Q867 A0A1I8Q836 Q7QEW8 A0A182X7Z9 A0A182HIB8 A0A182N1L0 B4PYC9 A0A0P8XL03 B3MVZ7 B3NV80 A0A1W4V242 B7Z139 A8JUR4 B4R2S8 A0A3B0J785 A0A182P2A7 M9PGW1 Q9VZ07 A0A182W502 B5DM22 A0A182HBS8 A8JUR5 A0A0M4EXV3 A0A3B0JC21 B4JL35 A0A0K8W6T2 D6WK33 B4L7I2 A0A2P8ZFZ3 A0A2M4CS53 A0A0Q9VZE0 A0A2M4CTK2 A0A232FJW3 A0A2M4CSA3 A0A0Q9VZF3 A0A2M4A7P2 A0A2M4A7L9 K7IWB7 E2APE2 E9IPB0 F4X2C7 A0A0C9PRJ8 E2BKQ4 A0A151XGA3
A0A212ETY8 A0A2H1WIV4 A0A023EXS2 A0A1B0D2V3 Q17J47 A0A1B0C902 A0A1Q3FAC7 A0A1Q3FAC1 A0A1L8DUQ5 A0A1Q3FAA0 A0A1L8DUQ1 B0WM84 A0A1L8DVG0 A0A1Q3FA95 A0A1L8DVH1 A0A1A9VIK3 A0A182G522 A0A1B0BHR2 A0A1B0A9Z5 A0A1A9WCC5 A0A1B0FQ22 A0A1I8MFF7 A0A1A9XC32 A0A0A1X335 W8BVT7 A0A0K8UF20 A0A1I8MFD8 A0A1I8MFE0 W8AWE6 W8BJ22 A0A034VN10 A0A1J1IPX6 A0A182IQU9 A0A0A1XEH2 A0A0A1WLD8 A0A0K8WLV1 A0A0K8TVC2 A0A084WPG0 A0A0L0BNH1 A0A1I8Q823 A0A182M7G2 A0A182R7I5 B4NES2 A0A336KQW8 A0A1I8Q864 A0A182QJ16 A0A182YQ66 A0A182V6E4 A0A182K7A1 A0A1I8Q867 A0A1I8Q836 Q7QEW8 A0A182X7Z9 A0A182HIB8 A0A182N1L0 B4PYC9 A0A0P8XL03 B3MVZ7 B3NV80 A0A1W4V242 B7Z139 A8JUR4 B4R2S8 A0A3B0J785 A0A182P2A7 M9PGW1 Q9VZ07 A0A182W502 B5DM22 A0A182HBS8 A8JUR5 A0A0M4EXV3 A0A3B0JC21 B4JL35 A0A0K8W6T2 D6WK33 B4L7I2 A0A2P8ZFZ3 A0A2M4CS53 A0A0Q9VZE0 A0A2M4CTK2 A0A232FJW3 A0A2M4CSA3 A0A0Q9VZF3 A0A2M4A7P2 A0A2M4A7L9 K7IWB7 E2APE2 E9IPB0 F4X2C7 A0A0C9PRJ8 E2BKQ4 A0A151XGA3
Ontologies
GO
PANTHER
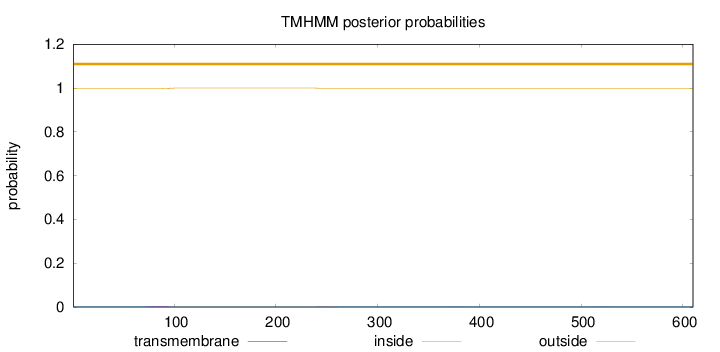
Topology
Length:
610
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0795000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00295
outside
1 - 610
Population Genetic Test Statistics
Pi
264.010336
Theta
219.042165
Tajima's D
0.390938
CLR
0.356821
CSRT
0.48307584620769
Interpretation
Uncertain
