Gene
KWMTBOMO01926
Annotation
PREDICTED:_reactive_oxygen_species_modulator_1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.07 Extracellular Reliability : 1.119 Mitochondrial Reliability : 1.429 Nuclear Reliability : 1.079
Sequence
CDS
ATGCCTGTACCGGGTGCTGTTTATCACAACCAAGGACCTTCGTGTTTTGATAAAATGAAAATGGGTTTTATGATTGGTTTTTGTGTAGGGATGGCTAGTGGAGGTCTTTTCGGCGGATTTACTGCATTAAGGTATGGAGCAAGGGGGAAAGAGTTAGTACATTCCGTTGGAAAAGTAATGCTTCAAGGTGGAGGCACATTTGGGACTTTTATGGCTATTGGAACTGGTATACGTTGTTGA
Protein
MPVPGAVYHNQGPSCFDKMKMGFMIGFCVGMASGGLFGGFTALRYGARGKELVHSVGKVMLQGGGTFGTFMAIGTGIRC
Summary
Uniprot
A0A194RCI2
A0A2H1WCC9
A0A194PKK8
A0A212ETV5
S4P3R4
A0A0L7LN58
+ More
A0A0K8TPW5 A0A1D2N8D2 A0A1W4XCY5 A0A1A9WE17 T1DGJ2 A0A2M4CTC3 A0A2M3ZAQ2 A0A2M4AK80 A0A2M4C628 A0A182MXD7 A0A182FV98 A0A1I8N4C4 A0A1B0FJL6 A0A1B0CH59 A0A0L0C2Y8 A0A1I8PGE7 A0A1L8EEK9 B4N516 B3M727 A0A182P1K1 A0A182V169 A0A182JV07 A0A182X023 Q7PZY4 A0A182HPQ7 A0A182R1X0 A0A182Q3B3 A0A182WD80 A0A182YRI7 Q2M1A8 B4GUD7 A0A067R7D0 A0A1W4VMZ3 A0A1Q3FGK0 B4IZ26 B4PCE1 B4HI62 B3NI03 B4QKL3 Q9VUM2 T1DJA5 Q16YX8 A0A023EBL5 U5EWW4 B4LGV0 A0A0L7QVM7 A0A195DCQ1 A0A195BCJ7 B0W567 A0A1L8DZP5 A0A2C9K734 B4NIB8 A0A0M4EM55 A0A1B0DBY8 A0A2J7PUT3 A0A195C668 T1JJU5 A0A131XQ08 A0A1E1WZ58 A0A2P8XMY8 A0A023GEC7 G3MNA2 A0A224YPX6 A0A131Z4F8 A0A0C9S9P8 G3MMY7 A0A2R7X948 N6TAD0 A0A1J1ILM2 T1FMP8 A0A1Z5LBR7 T1J6A0 A0A0K8RGL1 B7PAN4 B4L134 A0A293LWG7 T1JE58 A0A2R5LFW1 A0A1S3D2R5 A0A026WB97 A0A154P6K4 A0A0V0G727 A0A069DNQ7 A0A023F998 A0A023FMZ0 A0A0P4VUV7 R4G8H2 A0A2H5CUB5 A0A2A3ENF7 A0A088A1C8 V9IAY1 A0A0M8ZXV6 A0A1D1UXW8 E0VF23
A0A0K8TPW5 A0A1D2N8D2 A0A1W4XCY5 A0A1A9WE17 T1DGJ2 A0A2M4CTC3 A0A2M3ZAQ2 A0A2M4AK80 A0A2M4C628 A0A182MXD7 A0A182FV98 A0A1I8N4C4 A0A1B0FJL6 A0A1B0CH59 A0A0L0C2Y8 A0A1I8PGE7 A0A1L8EEK9 B4N516 B3M727 A0A182P1K1 A0A182V169 A0A182JV07 A0A182X023 Q7PZY4 A0A182HPQ7 A0A182R1X0 A0A182Q3B3 A0A182WD80 A0A182YRI7 Q2M1A8 B4GUD7 A0A067R7D0 A0A1W4VMZ3 A0A1Q3FGK0 B4IZ26 B4PCE1 B4HI62 B3NI03 B4QKL3 Q9VUM2 T1DJA5 Q16YX8 A0A023EBL5 U5EWW4 B4LGV0 A0A0L7QVM7 A0A195DCQ1 A0A195BCJ7 B0W567 A0A1L8DZP5 A0A2C9K734 B4NIB8 A0A0M4EM55 A0A1B0DBY8 A0A2J7PUT3 A0A195C668 T1JJU5 A0A131XQ08 A0A1E1WZ58 A0A2P8XMY8 A0A023GEC7 G3MNA2 A0A224YPX6 A0A131Z4F8 A0A0C9S9P8 G3MMY7 A0A2R7X948 N6TAD0 A0A1J1ILM2 T1FMP8 A0A1Z5LBR7 T1J6A0 A0A0K8RGL1 B7PAN4 B4L134 A0A293LWG7 T1JE58 A0A2R5LFW1 A0A1S3D2R5 A0A026WB97 A0A154P6K4 A0A0V0G727 A0A069DNQ7 A0A023F998 A0A023FMZ0 A0A0P4VUV7 R4G8H2 A0A2H5CUB5 A0A2A3ENF7 A0A088A1C8 V9IAY1 A0A0M8ZXV6 A0A1D1UXW8 E0VF23
Pubmed
26354079
22118469
23622113
26227816
26369729
27289101
+ More
25315136 26108605 17994087 12364791 14747013 17210077 25244985 15632085 24845553 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24330624 17510324 24945155 15562597 28049606 28503490 29403074 22216098 28797301 26830274 26131772 23537049 23254933 28528879 24508170 30249741 26334808 25474469 27129103 28889015 27649274 20566863
25315136 26108605 17994087 12364791 14747013 17210077 25244985 15632085 24845553 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24330624 17510324 24945155 15562597 28049606 28503490 29403074 22216098 28797301 26830274 26131772 23537049 23254933 28528879 24508170 30249741 26334808 25474469 27129103 28889015 27649274 20566863
EMBL
KQ460398
KPJ15179.1
ODYU01007708
SOQ50735.1
KQ459601
KPI93862.1
+ More
AGBW02012530 OWR44918.1 GAIX01011495 JAA81065.1 JTDY01000501 KOB76882.1 GDAI01001435 JAI16168.1 LJIJ01000148 ODN01512.1 GAMD01002756 JAA98834.1 GGFL01004392 MBW68570.1 GGFM01004858 MBW25609.1 GGFK01007836 MBW41157.1 GGFJ01011277 MBW60418.1 CCAG010020214 AJWK01012066 JRES01001072 KNC25779.1 GFDG01001647 JAV17152.1 CH964101 EDW79455.1 CH902618 EDV40892.1 AAAB01008986 EAA00196.2 APCN01003267 AXCN02001696 CH379069 EAL30667.1 CH479191 EDW26220.1 KK852691 KDR18366.1 GFDL01008368 JAV26677.1 CH916366 EDV97734.1 CM000159 EDW94851.1 CH480815 EDW41558.1 CH954178 EDV52089.1 CM000363 CM002912 EDX10524.1 KMY99717.1 AE014296 AY070706 AAF49654.1 AAL48177.1 GALA01000386 JAA94466.1 CH477506 EAT39849.1 GAPW01006736 GAPW01006735 GAPW01006734 JAC06863.1 GANO01000368 JAB59503.1 CH940647 EDW70565.1 KQ414725 KOC62678.1 KQ980989 KYN10658.1 KQ976528 KYM81919.1 DS231841 EDS34813.1 GFDF01002136 JAV11948.1 CH964272 EDW83700.1 CP012525 ALC44672.1 AJVK01030405 NEVH01021193 PNF20098.1 KQ978220 KYM96359.1 JH432094 GEFH01000363 JAP68218.1 GFAC01007132 JAT92056.1 PYGN01001685 PSN33356.1 GBBM01003219 JAC32199.1 JO843353 AEO34970.1 GFPF01004744 MAA15890.1 GEDV01002274 JAP86283.1 GBZX01003043 JAG89697.1 JO843238 AEO34855.1 KK862804 PTY28313.1 APGK01037114 KB740948 KB631899 ENN77229.1 ERL86998.1 CVRI01000055 CRL01141.1 AMQM01002972 KB095959 ESO09468.1 GFJQ02002133 JAW04837.1 JH431874 GADI01004194 JAA69614.1 ABJB010108293 ABJB010206369 ABJB010581446 ABJB010668641 DS672165 EEC03656.1 CH933809 EDW18191.1 GFWV01007559 MAA32289.1 JH432116 GGLE01004280 MBY08406.1 KK107293 QOIP01000005 EZA53367.1 RLU22779.1 KQ434819 KZC06974.1 GECL01002331 JAP03793.1 GBGD01003667 JAC85222.1 GBBI01000635 GEMB01006807 JAC18077.1 JAR96548.1 GBBK01001360 JAC23122.1 GDKW01001998 JAI54597.1 ACPB03006711 GAHY01000977 JAA76533.1 KY680285 AUH28159.1 KZ288215 PBC32782.1 JR037139 JR037141 AEY57605.1 AEY57606.1 KQ435803 KOX73157.1 BDGG01000002 GAU92542.1 DS235100 EEB11996.1
AGBW02012530 OWR44918.1 GAIX01011495 JAA81065.1 JTDY01000501 KOB76882.1 GDAI01001435 JAI16168.1 LJIJ01000148 ODN01512.1 GAMD01002756 JAA98834.1 GGFL01004392 MBW68570.1 GGFM01004858 MBW25609.1 GGFK01007836 MBW41157.1 GGFJ01011277 MBW60418.1 CCAG010020214 AJWK01012066 JRES01001072 KNC25779.1 GFDG01001647 JAV17152.1 CH964101 EDW79455.1 CH902618 EDV40892.1 AAAB01008986 EAA00196.2 APCN01003267 AXCN02001696 CH379069 EAL30667.1 CH479191 EDW26220.1 KK852691 KDR18366.1 GFDL01008368 JAV26677.1 CH916366 EDV97734.1 CM000159 EDW94851.1 CH480815 EDW41558.1 CH954178 EDV52089.1 CM000363 CM002912 EDX10524.1 KMY99717.1 AE014296 AY070706 AAF49654.1 AAL48177.1 GALA01000386 JAA94466.1 CH477506 EAT39849.1 GAPW01006736 GAPW01006735 GAPW01006734 JAC06863.1 GANO01000368 JAB59503.1 CH940647 EDW70565.1 KQ414725 KOC62678.1 KQ980989 KYN10658.1 KQ976528 KYM81919.1 DS231841 EDS34813.1 GFDF01002136 JAV11948.1 CH964272 EDW83700.1 CP012525 ALC44672.1 AJVK01030405 NEVH01021193 PNF20098.1 KQ978220 KYM96359.1 JH432094 GEFH01000363 JAP68218.1 GFAC01007132 JAT92056.1 PYGN01001685 PSN33356.1 GBBM01003219 JAC32199.1 JO843353 AEO34970.1 GFPF01004744 MAA15890.1 GEDV01002274 JAP86283.1 GBZX01003043 JAG89697.1 JO843238 AEO34855.1 KK862804 PTY28313.1 APGK01037114 KB740948 KB631899 ENN77229.1 ERL86998.1 CVRI01000055 CRL01141.1 AMQM01002972 KB095959 ESO09468.1 GFJQ02002133 JAW04837.1 JH431874 GADI01004194 JAA69614.1 ABJB010108293 ABJB010206369 ABJB010581446 ABJB010668641 DS672165 EEC03656.1 CH933809 EDW18191.1 GFWV01007559 MAA32289.1 JH432116 GGLE01004280 MBY08406.1 KK107293 QOIP01000005 EZA53367.1 RLU22779.1 KQ434819 KZC06974.1 GECL01002331 JAP03793.1 GBGD01003667 JAC85222.1 GBBI01000635 GEMB01006807 JAC18077.1 JAR96548.1 GBBK01001360 JAC23122.1 GDKW01001998 JAI54597.1 ACPB03006711 GAHY01000977 JAA76533.1 KY680285 AUH28159.1 KZ288215 PBC32782.1 JR037139 JR037141 AEY57605.1 AEY57606.1 KQ435803 KOX73157.1 BDGG01000002 GAU92542.1 DS235100 EEB11996.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000037510
UP000094527
UP000192223
+ More
UP000091820 UP000075884 UP000069272 UP000095301 UP000092444 UP000092461 UP000037069 UP000095300 UP000007798 UP000007801 UP000075885 UP000075903 UP000075881 UP000076407 UP000007062 UP000075840 UP000075900 UP000075886 UP000075920 UP000076408 UP000001819 UP000008744 UP000027135 UP000192221 UP000001070 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000008820 UP000008792 UP000053825 UP000078492 UP000078540 UP000002320 UP000076420 UP000092553 UP000092462 UP000235965 UP000078542 UP000245037 UP000019118 UP000030742 UP000183832 UP000015101 UP000001555 UP000009192 UP000079169 UP000053097 UP000279307 UP000076502 UP000015103 UP000242457 UP000005203 UP000053105 UP000186922 UP000009046
UP000091820 UP000075884 UP000069272 UP000095301 UP000092444 UP000092461 UP000037069 UP000095300 UP000007798 UP000007801 UP000075885 UP000075903 UP000075881 UP000076407 UP000007062 UP000075840 UP000075900 UP000075886 UP000075920 UP000076408 UP000001819 UP000008744 UP000027135 UP000192221 UP000001070 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000008820 UP000008792 UP000053825 UP000078492 UP000078540 UP000002320 UP000076420 UP000092553 UP000092462 UP000235965 UP000078542 UP000245037 UP000019118 UP000030742 UP000183832 UP000015101 UP000001555 UP000009192 UP000079169 UP000053097 UP000279307 UP000076502 UP000015103 UP000242457 UP000005203 UP000053105 UP000186922 UP000009046
PRIDE
Pfam
PF10247 Romo1
Interpro
IPR018450
Romo1/Mgr2
ProteinModelPortal
A0A194RCI2
A0A2H1WCC9
A0A194PKK8
A0A212ETV5
S4P3R4
A0A0L7LN58
+ More
A0A0K8TPW5 A0A1D2N8D2 A0A1W4XCY5 A0A1A9WE17 T1DGJ2 A0A2M4CTC3 A0A2M3ZAQ2 A0A2M4AK80 A0A2M4C628 A0A182MXD7 A0A182FV98 A0A1I8N4C4 A0A1B0FJL6 A0A1B0CH59 A0A0L0C2Y8 A0A1I8PGE7 A0A1L8EEK9 B4N516 B3M727 A0A182P1K1 A0A182V169 A0A182JV07 A0A182X023 Q7PZY4 A0A182HPQ7 A0A182R1X0 A0A182Q3B3 A0A182WD80 A0A182YRI7 Q2M1A8 B4GUD7 A0A067R7D0 A0A1W4VMZ3 A0A1Q3FGK0 B4IZ26 B4PCE1 B4HI62 B3NI03 B4QKL3 Q9VUM2 T1DJA5 Q16YX8 A0A023EBL5 U5EWW4 B4LGV0 A0A0L7QVM7 A0A195DCQ1 A0A195BCJ7 B0W567 A0A1L8DZP5 A0A2C9K734 B4NIB8 A0A0M4EM55 A0A1B0DBY8 A0A2J7PUT3 A0A195C668 T1JJU5 A0A131XQ08 A0A1E1WZ58 A0A2P8XMY8 A0A023GEC7 G3MNA2 A0A224YPX6 A0A131Z4F8 A0A0C9S9P8 G3MMY7 A0A2R7X948 N6TAD0 A0A1J1ILM2 T1FMP8 A0A1Z5LBR7 T1J6A0 A0A0K8RGL1 B7PAN4 B4L134 A0A293LWG7 T1JE58 A0A2R5LFW1 A0A1S3D2R5 A0A026WB97 A0A154P6K4 A0A0V0G727 A0A069DNQ7 A0A023F998 A0A023FMZ0 A0A0P4VUV7 R4G8H2 A0A2H5CUB5 A0A2A3ENF7 A0A088A1C8 V9IAY1 A0A0M8ZXV6 A0A1D1UXW8 E0VF23
A0A0K8TPW5 A0A1D2N8D2 A0A1W4XCY5 A0A1A9WE17 T1DGJ2 A0A2M4CTC3 A0A2M3ZAQ2 A0A2M4AK80 A0A2M4C628 A0A182MXD7 A0A182FV98 A0A1I8N4C4 A0A1B0FJL6 A0A1B0CH59 A0A0L0C2Y8 A0A1I8PGE7 A0A1L8EEK9 B4N516 B3M727 A0A182P1K1 A0A182V169 A0A182JV07 A0A182X023 Q7PZY4 A0A182HPQ7 A0A182R1X0 A0A182Q3B3 A0A182WD80 A0A182YRI7 Q2M1A8 B4GUD7 A0A067R7D0 A0A1W4VMZ3 A0A1Q3FGK0 B4IZ26 B4PCE1 B4HI62 B3NI03 B4QKL3 Q9VUM2 T1DJA5 Q16YX8 A0A023EBL5 U5EWW4 B4LGV0 A0A0L7QVM7 A0A195DCQ1 A0A195BCJ7 B0W567 A0A1L8DZP5 A0A2C9K734 B4NIB8 A0A0M4EM55 A0A1B0DBY8 A0A2J7PUT3 A0A195C668 T1JJU5 A0A131XQ08 A0A1E1WZ58 A0A2P8XMY8 A0A023GEC7 G3MNA2 A0A224YPX6 A0A131Z4F8 A0A0C9S9P8 G3MMY7 A0A2R7X948 N6TAD0 A0A1J1ILM2 T1FMP8 A0A1Z5LBR7 T1J6A0 A0A0K8RGL1 B7PAN4 B4L134 A0A293LWG7 T1JE58 A0A2R5LFW1 A0A1S3D2R5 A0A026WB97 A0A154P6K4 A0A0V0G727 A0A069DNQ7 A0A023F998 A0A023FMZ0 A0A0P4VUV7 R4G8H2 A0A2H5CUB5 A0A2A3ENF7 A0A088A1C8 V9IAY1 A0A0M8ZXV6 A0A1D1UXW8 E0VF23
Ontologies
PANTHER
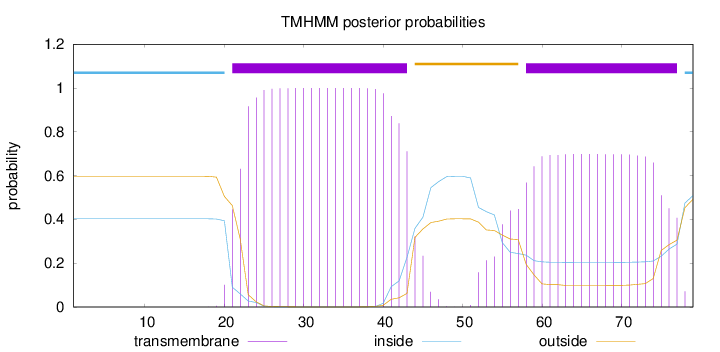
Topology
Length:
79
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
37.00969
Exp number, first 60 AAs:
25.86764
Total prob of N-in:
0.40274
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 57
TMhelix
58 - 77
inside
78 - 79
Population Genetic Test Statistics
Pi
10.487501
Theta
12.907487
Tajima's D
-0.592146
CLR
0
CSRT
0.223038848057597
Interpretation
Uncertain
