Gene
KWMTBOMO01924 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006214
Annotation
very_low-density_lipoprotein_receptor_isoform_1_precursor_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.123
Sequence
CDS
ATGGCTCGACACGCGTTTCTGTGTCTCGGTGTGTTATTAGTTGGTTCGCTGGTATGCAGTACTTCTCGTGTGAAGCGACAAGATGACGACGGAGCTGGAGATGAGCCGAACGCTGATCAGTTGTGCGACGGTAGACCCGCCGATGAATATTTCCGTCTCACCACCGAAGGTGACTGCCGCGATGTCGTCAGGTGCGACCAGGGCCTAGAGAACAGCGTCACCAGGCTCGCCTCAGTGCGCTGCCCCGGAGGTCTCGCCTTCGATATCGACCGTCAAACTTGTGATTGGAAAACGAATGTTAAGAATTGCGATCAAATAGAAAAGCCACGAAAAGTTTTACCAATCTTGAAAACGGACGAACCTATTTGTCCTGAAGGTAAATTGGCTTGTGGCAGCGGAGATTGCATTGAAAAAGAATTGTTCTGTAATGGTAAACCAGACTGCAAAGATGAATCCGATGAAAATGCTTGCACTGTCGAACTGGACCCAAATAGAGCGCCTGACTGTGATCCTAACCAGTGCGTCCTACCTGACTGTTTCTGTTCTGCTGACGGTACACGTATCCCAGGTGGAATTGAACCAAATCAAGTACCTCAAATGGTTACTATTACATTTAACGGTGCTGTGAATGTTGACAACATCGATTTATATGAACAAATATTCAACGGTAATCGTCATAATCCTAACGGTTGTCAAATTAAAGGAACGTTTTTCGTCTCACACAAGTATACTAACTATGCAGCTGTACAAGAATTACACCGCAAAGGACATGAAATTTCTGTATTTTCCATAACTCATAAAGATGATCCGCAATACTGGACGAGTGGTTCGTACGATGATTGGTTGGCTGAAATGGCCGGAGCTCGTCTTATCATTGAACGTTACGCAAATATCACTGACTCTTCTATAATTGGAGTTCGGGCGCCTTACTTACGTGTCGGTGGAAACAAACAATTCGAAATGATGGGTGATCAATACTTCGTGTATGATGCATCCATCACTGCTCCTCTTGGACGCGTCCCGATATGGCCTTACACATTGTATTTCCGCATGCCACATAAGTGTAACGGCAACGCCCACAACTGTCCGTCTAGAAGTCACCCCGTTTGGGAAATGGTCATGAATGAATTGGACAGAAGGGACGATCCCACATTTGATGAATCTCTACCAGGTTGCCACGTTGTAGACTCATGTTCAAACATCCAAAGTGGTGAACAATTTGGACGTCTTCTCCGACACAACTTCAACCGTCATTACACAACCAACCGCGCTCCCTTAGGCTTCCACTTCCACGCCTCCTGGCTTAAATCTAAAAAAGAATACAGAGATGAATTAATCAAGTTTATCGAAGAAATGTTGGAAAAGAACGATGTATACTTCACTTCACTAATTCAGGTTATTCAATGGATGCAAAACCCTACCGAATTATCATCACTCCGTGACTTCCAAGAATGGAAACAAGATAAATGTGACGTCAAAGGTCAACCTTTCTGTTCCCTGCCGAACGCCTGTCCACTAACAACTAGAGAGTTACCAGGAGAGACATTACGTCTTTTCACTTGTATGGAGTGTCCTAACAACTATCCATGGATCTTGGATCCTCAGGGCGAAGGATACAACGTTAAGAAGTGA
Protein
MARHAFLCLGVLLVGSLVCSTSRVKRQDDDGAGDEPNADQLCDGRPADEYFRLTTEGDCRDVVRCDQGLENSVTRLASVRCPGGLAFDIDRQTCDWKTNVKNCDQIEKPRKVLPILKTDEPICPEGKLACGSGDCIEKELFCNGKPDCKDESDENACTVELDPNRAPDCDPNQCVLPDCFCSADGTRIPGGIEPNQVPQMVTITFNGAVNVDNIDLYEQIFNGNRHNPNGCQIKGTFFVSHKYTNYAAVQELHRKGHEISVFSITHKDDPQYWTSGSYDDWLAEMAGARLIIERYANITDSSIIGVRAPYLRVGGNKQFEMMGDQYFVYDASITAPLGRVPIWPYTLYFRMPHKCNGNAHNCPSRSHPVWEMVMNELDRRDDPTFDESLPGCHVVDSCSNIQSGEQFGRLLRHNFNRHYTTNRAPLGFHFHASWLKSKKEYRDELIKFIEEMLEKNDVYFTSLIQVIQWMQNPTELSSLRDFQEWKQDKCDVKGQPFCSLPNACPLTTRELPGETLRLFTCMECPNNYPWILDPQGEGYNVKK
Summary
Similarity
Belongs to the universal ribosomal protein uS9 family.
Uniprot
Q1HQA6
H9J9M1
Q1HQA5
A0A2W1BWV5
A0A2A4K3P0
A0A1E1W4N0
+ More
A0A2H1WCQ4 Q1HQA4 S5Z6S1 A0A0G2RLP0 A0A2P0MAJ4 A0A194RD29 A0A2A4K3A7 A0A194PLI9 A0A0A7R7B5 S5YT69 B2DBL8 A0A0L7LN78 I4DM00 A0A212ETV0 A0A2U7LR78 A0A2R2PTF4 A0A1L2DBW3 A0A182JAH8 A0A0A9WUV8 A0A2J7PUN9 A0A182Q6G6 A0A2K8JSV4 A0A1S4F4K8 A0A023ETQ8 A0A1S3DAU6 Q17FD0 A0A182N2S3 A0A2M4AHJ5 A0A1B0CSR2 A0A182FB83 A0A182RXV8 A0A1J1HN62 A0A182UNW3 A0A182Y9I7 A0A2P8XHQ7 A0A336LPN1 A0A182HHM6 A0A182MBH9 A0A1Q3FZ89 A0A182WE18 A0A182PMW5 A0A067RGU7 A0A0P6GVX1 A0A1Q3FMZ7 A0A023F5Q3 A0A224XL71 A0A161TH04 A0A0C9QC03 A0A0F6PJH0 A0A088A1P7 A0A2R2PTF2 A0A0L7QVU3 A0A1B6HBE8 A0A182JAH7 A0A336M3L8 A0A1L2DBY0 A0A182GLF5 Q7PKC7 A0A3L8DH86 A0A182N2S2 A0A084WIH1 A0A1S4F4T2 A0A139WI93 A0A182FB84 Q17FD1 A0A182HHM5 A0A182RXV7 A0A2M4AHD3 W5JTL4 A0A182WE17 B0WLQ8 A0A1A9XK26 A0A1W4XB71 A0A1B0ANM8 A0A1Q3FN30 A7U8D0 A0A1A9ZBV8 A0A0T6B567 N6U849 A0A2J7PUR0 A0A336LSB4 J3JVM2 A0A1S3DC94 A0A2Z2Z237 W8B0I5 A0A2P1NQ38 A0A1J1HTI8 A0A0A1X909 A0A1B6CBX2 A0A1I8P760 Q06RG8 A0A0P6G7L7 A0A171AS08 X1WI58
A0A2H1WCQ4 Q1HQA4 S5Z6S1 A0A0G2RLP0 A0A2P0MAJ4 A0A194RD29 A0A2A4K3A7 A0A194PLI9 A0A0A7R7B5 S5YT69 B2DBL8 A0A0L7LN78 I4DM00 A0A212ETV0 A0A2U7LR78 A0A2R2PTF4 A0A1L2DBW3 A0A182JAH8 A0A0A9WUV8 A0A2J7PUN9 A0A182Q6G6 A0A2K8JSV4 A0A1S4F4K8 A0A023ETQ8 A0A1S3DAU6 Q17FD0 A0A182N2S3 A0A2M4AHJ5 A0A1B0CSR2 A0A182FB83 A0A182RXV8 A0A1J1HN62 A0A182UNW3 A0A182Y9I7 A0A2P8XHQ7 A0A336LPN1 A0A182HHM6 A0A182MBH9 A0A1Q3FZ89 A0A182WE18 A0A182PMW5 A0A067RGU7 A0A0P6GVX1 A0A1Q3FMZ7 A0A023F5Q3 A0A224XL71 A0A161TH04 A0A0C9QC03 A0A0F6PJH0 A0A088A1P7 A0A2R2PTF2 A0A0L7QVU3 A0A1B6HBE8 A0A182JAH7 A0A336M3L8 A0A1L2DBY0 A0A182GLF5 Q7PKC7 A0A3L8DH86 A0A182N2S2 A0A084WIH1 A0A1S4F4T2 A0A139WI93 A0A182FB84 Q17FD1 A0A182HHM5 A0A182RXV7 A0A2M4AHD3 W5JTL4 A0A182WE17 B0WLQ8 A0A1A9XK26 A0A1W4XB71 A0A1B0ANM8 A0A1Q3FN30 A7U8D0 A0A1A9ZBV8 A0A0T6B567 N6U849 A0A2J7PUR0 A0A336LSB4 J3JVM2 A0A1S3DC94 A0A2Z2Z237 W8B0I5 A0A2P1NQ38 A0A1J1HTI8 A0A0A1X909 A0A1B6CBX2 A0A1I8P760 Q06RG8 A0A0P6G7L7 A0A171AS08 X1WI58
Pubmed
19121390
28756777
23628857
26354079
18712529
26227816
+ More
22651552 22118469 27637332 25401762 26823975 24945155 17510324 25244985 29403074 24845553 25474469 24989071 26483478 12364791 14747013 17210077 30249741 24438588 18362917 19820115 20920257 23761445 23537049 22516182 24495485 25830018 18428030
22651552 22118469 27637332 25401762 26823975 24945155 17510324 25244985 29403074 24845553 25474469 24989071 26483478 12364791 14747013 17210077 30249741 24438588 18362917 19820115 20920257 23761445 23537049 22516182 24495485 25830018 18428030
EMBL
DQ443146
HM450149
ABF51235.1
ADO24153.1
BABH01005058
DQ443147
+ More
HM450150 ABF51236.1 ADO24154.1 KZ149895 PZC78821.1 NWSH01000193 PCG78528.1 GDQN01009134 JAT81920.1 ODYU01007709 SOQ50736.1 DQ443148 ABF51237.1 KC285591 KC295540 AGT28750.1 KP000852 AJG44547.1 KT781841 AOS49616.1 KQ460398 KPJ15180.1 PCG78529.1 KQ459601 KPI93863.1 KM598637 AJA30435.1 KC295541 AGT28749.1 AB264702 BAG30777.1 JTDY01000501 KOB76890.1 AK402318 BAM18940.1 AGBW02012530 OWR44920.1 KT781842 AOS49617.1 KR537804 ANA57444.1 KR537801 ANA57441.1 GBHO01031342 GBRD01011249 GDHC01004474 JAG12262.1 JAG54575.1 JAQ14155.1 NEVH01021193 PNF20055.1 AXCN02001087 KY031281 ATU83032.1 GAPW01000863 JAC12735.1 CH477271 EAT45315.1 GGFK01006944 MBW40265.1 AJWK01026427 AJWK01026428 AJWK01026429 AJWK01026430 AJWK01026431 CVRI01000012 CRK89496.1 PYGN01002090 PSN31537.1 UFQT01000099 UFQT01000360 SSX19870.1 SSX23524.1 APCN01002429 AXCM01001841 GFDL01002164 JAV32881.1 KK852691 KDR18357.1 GDIQ01040694 GDIQ01003925 JAN54043.1 GFDL01006074 JAV28971.1 GBBI01002214 JAC16498.1 GFTR01007074 JAW09352.1 GEMB01000725 JAS02412.1 GBYB01000764 JAG70531.1 KJ825887 AJQ20732.1 KR537805 ANA57445.1 KQ414725 KOC62666.1 GECU01035719 JAS71987.1 SSX19869.1 SSX23523.1 KR537802 ANA57442.1 JXUM01014451 JXUM01014452 JXUM01014453 KQ560404 KXJ82583.1 AAAB01008986 EAA43313.4 QOIP01000008 RLU19701.1 ATLV01023931 KE525347 KFB50015.1 KQ971342 KYB27624.1 EAT45314.1 GGFK01006870 MBW40191.1 ADMH02000120 ETN67707.1 DS231989 EDS30623.1 JXJN01000896 GFDL01006036 JAV29009.1 EU019712 ABU25224.1 LJIG01009733 KRT82489.1 APGK01045191 APGK01045192 APGK01045193 APGK01045194 KB741031 ENN74767.1 PNF20054.1 SSX19871.1 SSX23525.1 BT127290 AEE62252.1 KY681049 AUP42576.1 GAMC01014588 JAB91967.1 KY914479 AVP71459.1 CRK89497.1 GBXI01007134 JAD07158.1 GEDC01026362 JAS10936.1 DQ665309 ABI95429.1 GDIQ01048694 GDIQ01047199 GDIQ01040695 JAN46043.1 GEMB01000727 JAS02410.1 ABLF02025146
HM450150 ABF51236.1 ADO24154.1 KZ149895 PZC78821.1 NWSH01000193 PCG78528.1 GDQN01009134 JAT81920.1 ODYU01007709 SOQ50736.1 DQ443148 ABF51237.1 KC285591 KC295540 AGT28750.1 KP000852 AJG44547.1 KT781841 AOS49616.1 KQ460398 KPJ15180.1 PCG78529.1 KQ459601 KPI93863.1 KM598637 AJA30435.1 KC295541 AGT28749.1 AB264702 BAG30777.1 JTDY01000501 KOB76890.1 AK402318 BAM18940.1 AGBW02012530 OWR44920.1 KT781842 AOS49617.1 KR537804 ANA57444.1 KR537801 ANA57441.1 GBHO01031342 GBRD01011249 GDHC01004474 JAG12262.1 JAG54575.1 JAQ14155.1 NEVH01021193 PNF20055.1 AXCN02001087 KY031281 ATU83032.1 GAPW01000863 JAC12735.1 CH477271 EAT45315.1 GGFK01006944 MBW40265.1 AJWK01026427 AJWK01026428 AJWK01026429 AJWK01026430 AJWK01026431 CVRI01000012 CRK89496.1 PYGN01002090 PSN31537.1 UFQT01000099 UFQT01000360 SSX19870.1 SSX23524.1 APCN01002429 AXCM01001841 GFDL01002164 JAV32881.1 KK852691 KDR18357.1 GDIQ01040694 GDIQ01003925 JAN54043.1 GFDL01006074 JAV28971.1 GBBI01002214 JAC16498.1 GFTR01007074 JAW09352.1 GEMB01000725 JAS02412.1 GBYB01000764 JAG70531.1 KJ825887 AJQ20732.1 KR537805 ANA57445.1 KQ414725 KOC62666.1 GECU01035719 JAS71987.1 SSX19869.1 SSX23523.1 KR537802 ANA57442.1 JXUM01014451 JXUM01014452 JXUM01014453 KQ560404 KXJ82583.1 AAAB01008986 EAA43313.4 QOIP01000008 RLU19701.1 ATLV01023931 KE525347 KFB50015.1 KQ971342 KYB27624.1 EAT45314.1 GGFK01006870 MBW40191.1 ADMH02000120 ETN67707.1 DS231989 EDS30623.1 JXJN01000896 GFDL01006036 JAV29009.1 EU019712 ABU25224.1 LJIG01009733 KRT82489.1 APGK01045191 APGK01045192 APGK01045193 APGK01045194 KB741031 ENN74767.1 PNF20054.1 SSX19871.1 SSX23525.1 BT127290 AEE62252.1 KY681049 AUP42576.1 GAMC01014588 JAB91967.1 KY914479 AVP71459.1 CRK89497.1 GBXI01007134 JAD07158.1 GEDC01026362 JAS10936.1 DQ665309 ABI95429.1 GDIQ01048694 GDIQ01047199 GDIQ01040695 JAN46043.1 GEMB01000727 JAS02410.1 ABLF02025146
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000075880 UP000235965 UP000075886 UP000079169 UP000008820 UP000075884 UP000092461 UP000069272 UP000075900 UP000183832 UP000075903 UP000076408 UP000245037 UP000075840 UP000075883 UP000075920 UP000075885 UP000027135 UP000005203 UP000053825 UP000069940 UP000249989 UP000007062 UP000279307 UP000030765 UP000007266 UP000000673 UP000002320 UP000092443 UP000192223 UP000092460 UP000092445 UP000019118 UP000095300 UP000007819
UP000075880 UP000235965 UP000075886 UP000079169 UP000008820 UP000075884 UP000092461 UP000069272 UP000075900 UP000183832 UP000075903 UP000076408 UP000245037 UP000075840 UP000075883 UP000075920 UP000075885 UP000027135 UP000005203 UP000053825 UP000069940 UP000249989 UP000007062 UP000279307 UP000030765 UP000007266 UP000000673 UP000002320 UP000092443 UP000192223 UP000092460 UP000092445 UP000019118 UP000095300 UP000007819
Interpro
IPR002172
LDrepeatLR_classA_rpt
+ More
IPR036055 LDL_receptor-like_sf
IPR036508 Chitin-bd_dom_sf
IPR011330 Glyco_hydro/deAcase_b/a-brl
IPR023415 LDLR_class-A_CS
IPR002557 Chitin-bd_dom
IPR020574 Ribosomal_S9_CS
IPR020568 Ribosomal_S5_D2-typ_fold
IPR000754 Ribosomal_S9
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
IPR036055 LDL_receptor-like_sf
IPR036508 Chitin-bd_dom_sf
IPR011330 Glyco_hydro/deAcase_b/a-brl
IPR023415 LDLR_class-A_CS
IPR002557 Chitin-bd_dom
IPR020574 Ribosomal_S9_CS
IPR020568 Ribosomal_S5_D2-typ_fold
IPR000754 Ribosomal_S9
IPR014721 Ribosomal_S5_D2-typ_fold_subgr
Gene 3D
CDD
ProteinModelPortal
Q1HQA6
H9J9M1
Q1HQA5
A0A2W1BWV5
A0A2A4K3P0
A0A1E1W4N0
+ More
A0A2H1WCQ4 Q1HQA4 S5Z6S1 A0A0G2RLP0 A0A2P0MAJ4 A0A194RD29 A0A2A4K3A7 A0A194PLI9 A0A0A7R7B5 S5YT69 B2DBL8 A0A0L7LN78 I4DM00 A0A212ETV0 A0A2U7LR78 A0A2R2PTF4 A0A1L2DBW3 A0A182JAH8 A0A0A9WUV8 A0A2J7PUN9 A0A182Q6G6 A0A2K8JSV4 A0A1S4F4K8 A0A023ETQ8 A0A1S3DAU6 Q17FD0 A0A182N2S3 A0A2M4AHJ5 A0A1B0CSR2 A0A182FB83 A0A182RXV8 A0A1J1HN62 A0A182UNW3 A0A182Y9I7 A0A2P8XHQ7 A0A336LPN1 A0A182HHM6 A0A182MBH9 A0A1Q3FZ89 A0A182WE18 A0A182PMW5 A0A067RGU7 A0A0P6GVX1 A0A1Q3FMZ7 A0A023F5Q3 A0A224XL71 A0A161TH04 A0A0C9QC03 A0A0F6PJH0 A0A088A1P7 A0A2R2PTF2 A0A0L7QVU3 A0A1B6HBE8 A0A182JAH7 A0A336M3L8 A0A1L2DBY0 A0A182GLF5 Q7PKC7 A0A3L8DH86 A0A182N2S2 A0A084WIH1 A0A1S4F4T2 A0A139WI93 A0A182FB84 Q17FD1 A0A182HHM5 A0A182RXV7 A0A2M4AHD3 W5JTL4 A0A182WE17 B0WLQ8 A0A1A9XK26 A0A1W4XB71 A0A1B0ANM8 A0A1Q3FN30 A7U8D0 A0A1A9ZBV8 A0A0T6B567 N6U849 A0A2J7PUR0 A0A336LSB4 J3JVM2 A0A1S3DC94 A0A2Z2Z237 W8B0I5 A0A2P1NQ38 A0A1J1HTI8 A0A0A1X909 A0A1B6CBX2 A0A1I8P760 Q06RG8 A0A0P6G7L7 A0A171AS08 X1WI58
A0A2H1WCQ4 Q1HQA4 S5Z6S1 A0A0G2RLP0 A0A2P0MAJ4 A0A194RD29 A0A2A4K3A7 A0A194PLI9 A0A0A7R7B5 S5YT69 B2DBL8 A0A0L7LN78 I4DM00 A0A212ETV0 A0A2U7LR78 A0A2R2PTF4 A0A1L2DBW3 A0A182JAH8 A0A0A9WUV8 A0A2J7PUN9 A0A182Q6G6 A0A2K8JSV4 A0A1S4F4K8 A0A023ETQ8 A0A1S3DAU6 Q17FD0 A0A182N2S3 A0A2M4AHJ5 A0A1B0CSR2 A0A182FB83 A0A182RXV8 A0A1J1HN62 A0A182UNW3 A0A182Y9I7 A0A2P8XHQ7 A0A336LPN1 A0A182HHM6 A0A182MBH9 A0A1Q3FZ89 A0A182WE18 A0A182PMW5 A0A067RGU7 A0A0P6GVX1 A0A1Q3FMZ7 A0A023F5Q3 A0A224XL71 A0A161TH04 A0A0C9QC03 A0A0F6PJH0 A0A088A1P7 A0A2R2PTF2 A0A0L7QVU3 A0A1B6HBE8 A0A182JAH7 A0A336M3L8 A0A1L2DBY0 A0A182GLF5 Q7PKC7 A0A3L8DH86 A0A182N2S2 A0A084WIH1 A0A1S4F4T2 A0A139WI93 A0A182FB84 Q17FD1 A0A182HHM5 A0A182RXV7 A0A2M4AHD3 W5JTL4 A0A182WE17 B0WLQ8 A0A1A9XK26 A0A1W4XB71 A0A1B0ANM8 A0A1Q3FN30 A7U8D0 A0A1A9ZBV8 A0A0T6B567 N6U849 A0A2J7PUR0 A0A336LSB4 J3JVM2 A0A1S3DC94 A0A2Z2Z237 W8B0I5 A0A2P1NQ38 A0A1J1HTI8 A0A0A1X909 A0A1B6CBX2 A0A1I8P760 Q06RG8 A0A0P6G7L7 A0A171AS08 X1WI58
PDB
5ZNT
E-value=1.02297e-150,
Score=1369
Ontologies
GO
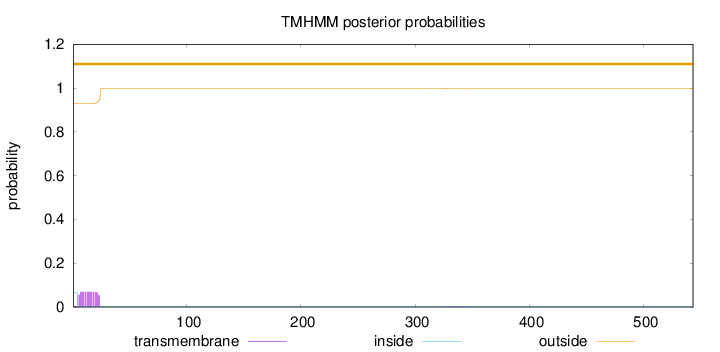
Topology
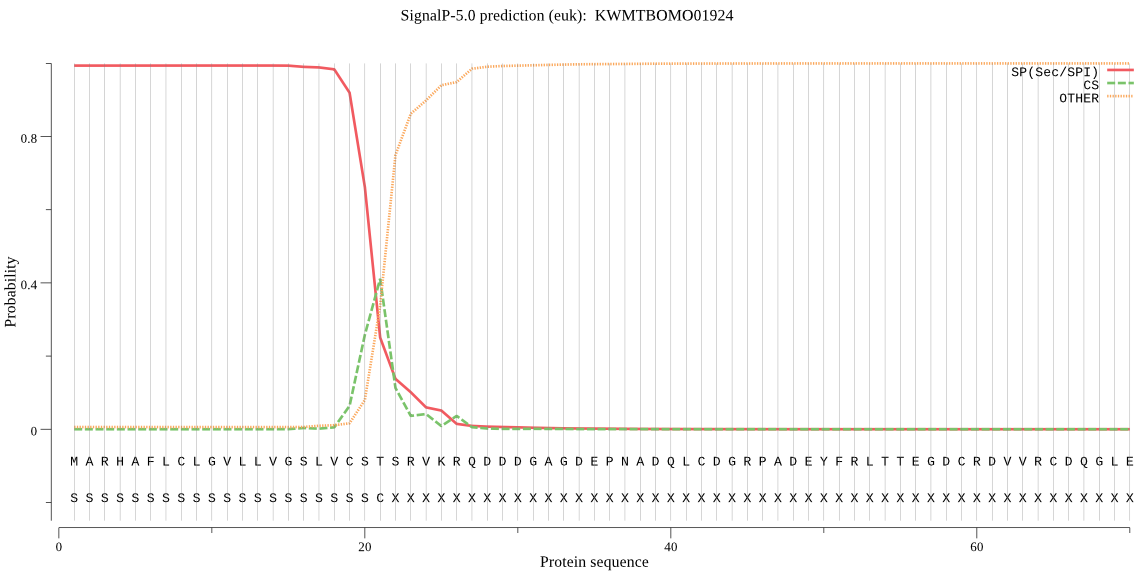
SignalP
Position: 1 - 21,
Likelihood: 0.993953
Length:
543
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.3986
Exp number, first 60 AAs:
1.33833
Total prob of N-in:
0.06962
outside
1 - 543
Population Genetic Test Statistics
Pi
294.796612
Theta
226.787756
Tajima's D
1.221315
CLR
0
CSRT
0.726113694315284
Interpretation
Uncertain
