Gene
KWMTBOMO01923 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006213
Annotation
PREDICTED:_uncharacterized_protein_LOC106143014_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.312
Sequence
CDS
ATGGCGCGCTACGCCCGTGTCGCTACTCTGGCCGCGTGCCTCTTGTTCGCCTGCGCGCTTGCTGATGGTCACCGATGGCGGCGACAAGCTGACGAGACGGCAAAGAAAGATGAAAGCTTGGAACAAGAACTATGCAAGGACAAGGACGCCGGCGAATGGTTCCGGCTGGTGGCCGGCGAGGGCGACAACTGTCGCGACGTCATCCAGTGTACTGCCTCGGGAATACAAGCTATACGTTGCCCGGCCGGTTTGTTCTTCGATATTGAGAAACAGACCTGCGATTGGAAAGATGCTGTGAAAAATTGTAAGCTCAAGAACAAGGAGCGTAAAATAAAGCCTCTCTTGTACACTGAAGAACCACTCTGTCAAGACGGCTTCCTCGCTTGCGGCGACTCTACTTGCATTGAACGCGGTCTTTTCTGTAACGGCGAAAAGGACTGTGGCGATGGATCTGATGAAAATTCTTGTGATATTGACAACGACCCAAATAGAGCTCCGCCATGCGATTCCTCGCAGTGTGTCCTTCCTGACTGCTTCTGCTCCGAGGACGGCACAGTGATCCCCGGCGACTTACCCGCCAGAGATGTCCCTCAAATGATCACTATCACCTTCGATGATGCGATTAACAATAACAACATTGAATTGTACAAAGAGATATTTAACGGCAAGCGCAAAAACCCAAACGGTTGCGATATCAAAGCCACTTATTTTGTCTCGCACAAGTACACTAACTATTCTGCCGTCCAAGAAACTCACCGCAAGGGTCACGAAATCGCAGTTCATTCTATTACGCACAATGACGATGAACGTTTTTGGAGCAACGCTACTGTCGATGATTGGGGCAAGGAAATGGCCGGTATGAGAGTTATTATTGAAAAATTCTCTAACATTACTGACAACAGCGTTGTAGGAGTTCGCGCACCTTATCTACGTGTAGGTGGCAATAATCAGTTCACCATGATGGAAGAACAGGCATTCTTATACGACAGCACTATTACAGCACCTTTATCAAACCCCCCTCTTTGGCCTTACACCATGTACTTTAGGATGCCCCACCGATGCCACGGAAACCTTCAGAGTTGCCCAACCAGGAGCCACGCAGTATGGGAAATGGTAATGAACGAGCTTGACAGACGTGAGGACCCAAGCAACGACGAATATTTACCCGGATGTGCCATGGTAGATTCTTGCTCTAACATTCTTACCGGTGATCAATTTTATAACTTCCTCAACCACAACTTTGACCGTCATTACGAACAGAACCGAGCACCTTTAGGACTTTATTTCCATGCAGCTTGGTTGAAAAACAACCCTGAATTTTTGGAAGCTTTCTTGTATTGGATAGACGAGATCCTTCAAAGCCACAACGACGTTTACTTCGTTACCATGACACAAGTGATCCAATGGGTACAAAACCCACGTACCGTGACTGAAGCCAAGAACTTCGAGCCGTGGAGAGAAAAGTGCTCCGTCGAAGGAAACCCTGCATGCTGGGTACCTCATTCCTGCAAGCTCACTTCAAAGGAAGTTCCCGGAGAAACCATCAATTTACAAACTTGTTTGAGATGCCCAGTCAACTACCCCTGGTTAAACGACCCCACGGGCGATGGCCACTATTAA
Protein
MARYARVATLAACLLFACALADGHRWRRQADETAKKDESLEQELCKDKDAGEWFRLVAGEGDNCRDVIQCTASGIQAIRCPAGLFFDIEKQTCDWKDAVKNCKLKNKERKIKPLLYTEEPLCQDGFLACGDSTCIERGLFCNGEKDCGDGSDENSCDIDNDPNRAPPCDSSQCVLPDCFCSEDGTVIPGDLPARDVPQMITITFDDAINNNNIELYKEIFNGKRKNPNGCDIKATYFVSHKYTNYSAVQETHRKGHEIAVHSITHNDDERFWSNATVDDWGKEMAGMRVIIEKFSNITDNSVVGVRAPYLRVGGNNQFTMMEEQAFLYDSTITAPLSNPPLWPYTMYFRMPHRCHGNLQSCPTRSHAVWEMVMNELDRREDPSNDEYLPGCAMVDSCSNILTGDQFYNFLNHNFDRHYEQNRAPLGLYFHAAWLKNNPEFLEAFLYWIDEILQSHNDVYFVTMTQVIQWVQNPRTVTEAKNFEPWREKCSVEGNPACWVPHSCKLTSKEVPGETINLQTCLRCPVNYPWLNDPTGDGHY
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M10A family.
Uniprot
H9J9M0
A0A2H1VHW8
I4DID1
I4DM33
F8SU33
A0A2A4K2Q6
+ More
D2WPC4 A0A0G2RLT9 B6CMG8 A0A0G3BDT6 A0A194RGX3 A0A218N0E9 D2K8M8 C4P7I6 A0A0L7LHC4 A0A182FZQ4 A0A182V2M0 A0A182WE16 A0A182XU93 A0A182L576 Q7PZI8 A0A182II05 A0A182S4K6 A0A1L8E3B3 A0A182Y9I6 A0A1B0CSR1 A0A084WIH3 A0A182JAG2 A0A346T6Q3 Q17FC9 A0A023EWC8 A0A182QAI1 A0A182GLF6 A0A336M566 A0A0F6PJR2 A0A182NZ11 A0A336LT18 A0A385P5D2 A7U8C9 A0A1I8PNX9 A0A0L0C8C8 A0A1W4UT92 B3M6J6 A0A1B6LFX8 B4N3M5 A0A0A1WHN2 B4KZA3 A0A0K8USM9 A0A2J7PUQ8 A0A2P8XDE0 A0A2R2PTG7 E2B171 R4G8G6 A0A3B0J9K9 Q29DE2 A0A2K8JWS3 A0A182MHG4 A0A1I8MBD5 A0A0J7L4R5 A0A1A9WH92 A0A2Z2YZJ3 A0A1B6ESY9 A0A0M4EC32 A0A067R7C1 A0A1S3DAQ8 A0A158NHE0 A0A212ETZ0 A0A1B6E5M7 B4LD22 A0A1B6LRM1 Q86P23 A0A088A1P6 B3NIH8 Q9VW34 A0A224XLA4 B4PFY9 A0A2A3EP13 B4IY40 B4QQX6 A0A195AZF0 A0A1J1ISL4 A0A195EE01 A0A195FYC2 A0A2J7PUR1 A0A1B6GS71 A0A0A9VWL5 J3JVK8 U4U9L0 A0A0K8SZS3 A0A195D5T8 A0A0T6B504 A0A026WT83 A0A0E3M394 A0A3L8DG03 A0A0C9RCT3 A0A1Y1MEQ4 A0A2S2QCD2 A0A2Z4N531 A0A232ESB3 J9JNW2
D2WPC4 A0A0G2RLT9 B6CMG8 A0A0G3BDT6 A0A194RGX3 A0A218N0E9 D2K8M8 C4P7I6 A0A0L7LHC4 A0A182FZQ4 A0A182V2M0 A0A182WE16 A0A182XU93 A0A182L576 Q7PZI8 A0A182II05 A0A182S4K6 A0A1L8E3B3 A0A182Y9I6 A0A1B0CSR1 A0A084WIH3 A0A182JAG2 A0A346T6Q3 Q17FC9 A0A023EWC8 A0A182QAI1 A0A182GLF6 A0A336M566 A0A0F6PJR2 A0A182NZ11 A0A336LT18 A0A385P5D2 A7U8C9 A0A1I8PNX9 A0A0L0C8C8 A0A1W4UT92 B3M6J6 A0A1B6LFX8 B4N3M5 A0A0A1WHN2 B4KZA3 A0A0K8USM9 A0A2J7PUQ8 A0A2P8XDE0 A0A2R2PTG7 E2B171 R4G8G6 A0A3B0J9K9 Q29DE2 A0A2K8JWS3 A0A182MHG4 A0A1I8MBD5 A0A0J7L4R5 A0A1A9WH92 A0A2Z2YZJ3 A0A1B6ESY9 A0A0M4EC32 A0A067R7C1 A0A1S3DAQ8 A0A158NHE0 A0A212ETZ0 A0A1B6E5M7 B4LD22 A0A1B6LRM1 Q86P23 A0A088A1P6 B3NIH8 Q9VW34 A0A224XLA4 B4PFY9 A0A2A3EP13 B4IY40 B4QQX6 A0A195AZF0 A0A1J1ISL4 A0A195EE01 A0A195FYC2 A0A2J7PUR1 A0A1B6GS71 A0A0A9VWL5 J3JVK8 U4U9L0 A0A0K8SZS3 A0A195D5T8 A0A0T6B504 A0A026WT83 A0A0E3M394 A0A3L8DG03 A0A0C9RCT3 A0A1Y1MEQ4 A0A2S2QCD2 A0A2Z4N531 A0A232ESB3 J9JNW2
Pubmed
19121390
30755482
22651552
26354079
20032185
28756777
+ More
18760362 26227816 20966253 12364791 14747013 17210077 25244985 24438588 17510324 24945155 26483478 24989071 18362917 19820115 26108605 17994087 25830018 29403074 27637332 20798317 15632085 25315136 24845553 21347285 22118469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 25401762 22516182 23537049 26823975 24508170 30249741 28004739 28648823
18760362 26227816 20966253 12364791 14747013 17210077 25244985 24438588 17510324 24945155 26483478 24989071 18362917 19820115 26108605 17994087 25830018 29403074 27637332 20798317 15632085 25315136 24845553 21347285 22118469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 22936249 25401762 22516182 23537049 26823975 24508170 30249741 28004739 28648823
EMBL
BABH01005057
BABH01005058
ODYU01002642
SOQ40419.1
AK401049
KQ459601
+ More
BAM17671.1 KPI93864.1 AK402351 BAM18973.1 HQ680620 AEI30868.1 NWSH01000193 PCG78527.1 GQ411189 KZ149895 ADB43610.1 PZC78822.1 KP000854 AJG44549.1 EU325568 ACB54958.1 KP348012 AKJ26157.1 KQ460398 KPJ15181.1 KY348778 ASF79656.1 GU188855 ACZ71481.1 FJ899541 ACQ73746.2 JTDY01001192 KOB74596.1 AAAB01008986 EAA00275.3 APCN01002429 GFDF01000873 JAV13211.1 AJWK01026423 AJWK01026424 AJWK01026425 ATLV01023931 ATLV01023932 KE525347 KFB50017.1 MF594382 AXU05961.1 CH477271 EAT45316.1 GAPW01000884 JAC12714.1 AXCN02001087 JXUM01014461 JXUM01014462 JXUM01014463 KQ560404 KXJ82584.1 UFQT01000360 SSX23527.1 KJ825888 AJQ20733.1 UFQT01000099 SSX19873.1 MG548738 AYA83838.1 EU019711 KQ971342 ABU25223.1 EFA03956.1 JRES01000760 KNC28526.1 CH902618 EDV40845.1 GEBQ01017427 JAT22550.1 CH964095 EDW79230.1 GBXI01015940 JAC98351.1 CH933809 EDW18929.1 GDHF01022796 JAI29518.1 NEVH01021193 PNF20067.1 PYGN01002763 PSN30003.1 KR537803 ANA57443.1 GL444799 EFN60563.1 ACPB03010253 GAHY01001007 JAA76503.1 OUUW01000002 SPP76632.1 CH379070 EAL30472.1 KY031116 ATU82867.1 AXCM01003562 LBMM01000736 KMQ97606.1 KY681047 AUP42574.1 GECZ01028728 JAS41041.1 CP012525 ALC45141.1 KK852691 KDR18356.1 ADTU01015712 AGBW02012530 OWR44921.1 GEDC01004080 JAS33218.1 CH940647 EDW69903.1 GEBQ01013723 JAT26254.1 BT003524 AAO39528.1 CH954178 EDV52403.1 AE014296 BT058041 AAF49119.2 ACM78483.1 AGB94755.1 GFTR01007044 JAW09382.1 CM000159 EDW95286.2 KZ288215 PBC32771.1 CH916366 EDV97583.1 CM000363 CM002912 EDX11114.1 KMZ00594.1 KQ976694 KYM77618.1 CVRI01000059 CRL03221.1 KQ979039 KYN23455.1 KQ981208 KYN44834.1 PNF20069.1 GECZ01004494 JAS65275.1 GBHO01045016 JAF98587.1 APGK01045180 APGK01045181 BT127276 KB741031 AEE62238.1 ENN74766.1 KB631842 ERL86620.1 GBRD01007059 GDHC01005326 JAG58762.1 JAQ13303.1 KQ976870 KYN07799.1 LJIG01009733 KRT82490.1 KK107109 EZA59222.1 KP271171 AKA64465.1 QOIP01000008 RLU19231.1 GBYB01014384 JAG84151.1 GEZM01033608 JAV84214.1 GGMS01006202 MBY75405.1 MF612049 AWX65381.1 NNAY01002467 OXU21232.1 ABLF02025147
BAM17671.1 KPI93864.1 AK402351 BAM18973.1 HQ680620 AEI30868.1 NWSH01000193 PCG78527.1 GQ411189 KZ149895 ADB43610.1 PZC78822.1 KP000854 AJG44549.1 EU325568 ACB54958.1 KP348012 AKJ26157.1 KQ460398 KPJ15181.1 KY348778 ASF79656.1 GU188855 ACZ71481.1 FJ899541 ACQ73746.2 JTDY01001192 KOB74596.1 AAAB01008986 EAA00275.3 APCN01002429 GFDF01000873 JAV13211.1 AJWK01026423 AJWK01026424 AJWK01026425 ATLV01023931 ATLV01023932 KE525347 KFB50017.1 MF594382 AXU05961.1 CH477271 EAT45316.1 GAPW01000884 JAC12714.1 AXCN02001087 JXUM01014461 JXUM01014462 JXUM01014463 KQ560404 KXJ82584.1 UFQT01000360 SSX23527.1 KJ825888 AJQ20733.1 UFQT01000099 SSX19873.1 MG548738 AYA83838.1 EU019711 KQ971342 ABU25223.1 EFA03956.1 JRES01000760 KNC28526.1 CH902618 EDV40845.1 GEBQ01017427 JAT22550.1 CH964095 EDW79230.1 GBXI01015940 JAC98351.1 CH933809 EDW18929.1 GDHF01022796 JAI29518.1 NEVH01021193 PNF20067.1 PYGN01002763 PSN30003.1 KR537803 ANA57443.1 GL444799 EFN60563.1 ACPB03010253 GAHY01001007 JAA76503.1 OUUW01000002 SPP76632.1 CH379070 EAL30472.1 KY031116 ATU82867.1 AXCM01003562 LBMM01000736 KMQ97606.1 KY681047 AUP42574.1 GECZ01028728 JAS41041.1 CP012525 ALC45141.1 KK852691 KDR18356.1 ADTU01015712 AGBW02012530 OWR44921.1 GEDC01004080 JAS33218.1 CH940647 EDW69903.1 GEBQ01013723 JAT26254.1 BT003524 AAO39528.1 CH954178 EDV52403.1 AE014296 BT058041 AAF49119.2 ACM78483.1 AGB94755.1 GFTR01007044 JAW09382.1 CM000159 EDW95286.2 KZ288215 PBC32771.1 CH916366 EDV97583.1 CM000363 CM002912 EDX11114.1 KMZ00594.1 KQ976694 KYM77618.1 CVRI01000059 CRL03221.1 KQ979039 KYN23455.1 KQ981208 KYN44834.1 PNF20069.1 GECZ01004494 JAS65275.1 GBHO01045016 JAF98587.1 APGK01045180 APGK01045181 BT127276 KB741031 AEE62238.1 ENN74766.1 KB631842 ERL86620.1 GBRD01007059 GDHC01005326 JAG58762.1 JAQ13303.1 KQ976870 KYN07799.1 LJIG01009733 KRT82490.1 KK107109 EZA59222.1 KP271171 AKA64465.1 QOIP01000008 RLU19231.1 GBYB01014384 JAG84151.1 GEZM01033608 JAV84214.1 GGMS01006202 MBY75405.1 MF612049 AWX65381.1 NNAY01002467 OXU21232.1 ABLF02025147
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000037510
UP000069272
+ More
UP000075903 UP000075920 UP000076407 UP000075882 UP000007062 UP000075840 UP000075900 UP000076408 UP000092461 UP000030765 UP000075880 UP000008820 UP000075886 UP000069940 UP000249989 UP000075884 UP000007266 UP000095300 UP000037069 UP000192221 UP000007801 UP000007798 UP000009192 UP000235965 UP000245037 UP000000311 UP000015103 UP000268350 UP000001819 UP000075883 UP000095301 UP000036403 UP000091820 UP000092553 UP000027135 UP000079169 UP000005205 UP000007151 UP000008792 UP000005203 UP000008711 UP000000803 UP000002282 UP000242457 UP000001070 UP000000304 UP000078540 UP000183832 UP000078492 UP000078541 UP000019118 UP000030742 UP000078542 UP000053097 UP000279307 UP000215335 UP000007819
UP000075903 UP000075920 UP000076407 UP000075882 UP000007062 UP000075840 UP000075900 UP000076408 UP000092461 UP000030765 UP000075880 UP000008820 UP000075886 UP000069940 UP000249989 UP000075884 UP000007266 UP000095300 UP000037069 UP000192221 UP000007801 UP000007798 UP000009192 UP000235965 UP000245037 UP000000311 UP000015103 UP000268350 UP000001819 UP000075883 UP000095301 UP000036403 UP000091820 UP000092553 UP000027135 UP000079169 UP000005205 UP000007151 UP000008792 UP000005203 UP000008711 UP000000803 UP000002282 UP000242457 UP000001070 UP000000304 UP000078540 UP000183832 UP000078492 UP000078541 UP000019118 UP000030742 UP000078542 UP000053097 UP000279307 UP000215335 UP000007819
Pfam
Interpro
IPR002172
LDrepeatLR_classA_rpt
+ More
IPR036055 LDL_receptor-like_sf
IPR036508 Chitin-bd_dom_sf
IPR002509 NODB_dom
IPR011330 Glyco_hydro/deAcase_b/a-brl
IPR023415 LDLR_class-A_CS
IPR002557 Chitin-bd_dom
IPR000585 Hemopexin-like_dom
IPR001818 Pept_M10_metallopeptidase
IPR011701 MFS
IPR018487 Hemopexin-like_repeat
IPR036259 MFS_trans_sf
IPR036365 PGBD-like_sf
IPR020846 MFS_dom
IPR024079 MetalloPept_cat_dom_sf
IPR036375 Hemopexin-like_dom_sf
IPR006026 Peptidase_Metallo
IPR033739 M10A_MMP
IPR021190 Pept_M10A
IPR036055 LDL_receptor-like_sf
IPR036508 Chitin-bd_dom_sf
IPR002509 NODB_dom
IPR011330 Glyco_hydro/deAcase_b/a-brl
IPR023415 LDLR_class-A_CS
IPR002557 Chitin-bd_dom
IPR000585 Hemopexin-like_dom
IPR001818 Pept_M10_metallopeptidase
IPR011701 MFS
IPR018487 Hemopexin-like_repeat
IPR036259 MFS_trans_sf
IPR036365 PGBD-like_sf
IPR020846 MFS_dom
IPR024079 MetalloPept_cat_dom_sf
IPR036375 Hemopexin-like_dom_sf
IPR006026 Peptidase_Metallo
IPR033739 M10A_MMP
IPR021190 Pept_M10A
SUPFAM
Gene 3D
ProteinModelPortal
H9J9M0
A0A2H1VHW8
I4DID1
I4DM33
F8SU33
A0A2A4K2Q6
+ More
D2WPC4 A0A0G2RLT9 B6CMG8 A0A0G3BDT6 A0A194RGX3 A0A218N0E9 D2K8M8 C4P7I6 A0A0L7LHC4 A0A182FZQ4 A0A182V2M0 A0A182WE16 A0A182XU93 A0A182L576 Q7PZI8 A0A182II05 A0A182S4K6 A0A1L8E3B3 A0A182Y9I6 A0A1B0CSR1 A0A084WIH3 A0A182JAG2 A0A346T6Q3 Q17FC9 A0A023EWC8 A0A182QAI1 A0A182GLF6 A0A336M566 A0A0F6PJR2 A0A182NZ11 A0A336LT18 A0A385P5D2 A7U8C9 A0A1I8PNX9 A0A0L0C8C8 A0A1W4UT92 B3M6J6 A0A1B6LFX8 B4N3M5 A0A0A1WHN2 B4KZA3 A0A0K8USM9 A0A2J7PUQ8 A0A2P8XDE0 A0A2R2PTG7 E2B171 R4G8G6 A0A3B0J9K9 Q29DE2 A0A2K8JWS3 A0A182MHG4 A0A1I8MBD5 A0A0J7L4R5 A0A1A9WH92 A0A2Z2YZJ3 A0A1B6ESY9 A0A0M4EC32 A0A067R7C1 A0A1S3DAQ8 A0A158NHE0 A0A212ETZ0 A0A1B6E5M7 B4LD22 A0A1B6LRM1 Q86P23 A0A088A1P6 B3NIH8 Q9VW34 A0A224XLA4 B4PFY9 A0A2A3EP13 B4IY40 B4QQX6 A0A195AZF0 A0A1J1ISL4 A0A195EE01 A0A195FYC2 A0A2J7PUR1 A0A1B6GS71 A0A0A9VWL5 J3JVK8 U4U9L0 A0A0K8SZS3 A0A195D5T8 A0A0T6B504 A0A026WT83 A0A0E3M394 A0A3L8DG03 A0A0C9RCT3 A0A1Y1MEQ4 A0A2S2QCD2 A0A2Z4N531 A0A232ESB3 J9JNW2
D2WPC4 A0A0G2RLT9 B6CMG8 A0A0G3BDT6 A0A194RGX3 A0A218N0E9 D2K8M8 C4P7I6 A0A0L7LHC4 A0A182FZQ4 A0A182V2M0 A0A182WE16 A0A182XU93 A0A182L576 Q7PZI8 A0A182II05 A0A182S4K6 A0A1L8E3B3 A0A182Y9I6 A0A1B0CSR1 A0A084WIH3 A0A182JAG2 A0A346T6Q3 Q17FC9 A0A023EWC8 A0A182QAI1 A0A182GLF6 A0A336M566 A0A0F6PJR2 A0A182NZ11 A0A336LT18 A0A385P5D2 A7U8C9 A0A1I8PNX9 A0A0L0C8C8 A0A1W4UT92 B3M6J6 A0A1B6LFX8 B4N3M5 A0A0A1WHN2 B4KZA3 A0A0K8USM9 A0A2J7PUQ8 A0A2P8XDE0 A0A2R2PTG7 E2B171 R4G8G6 A0A3B0J9K9 Q29DE2 A0A2K8JWS3 A0A182MHG4 A0A1I8MBD5 A0A0J7L4R5 A0A1A9WH92 A0A2Z2YZJ3 A0A1B6ESY9 A0A0M4EC32 A0A067R7C1 A0A1S3DAQ8 A0A158NHE0 A0A212ETZ0 A0A1B6E5M7 B4LD22 A0A1B6LRM1 Q86P23 A0A088A1P6 B3NIH8 Q9VW34 A0A224XLA4 B4PFY9 A0A2A3EP13 B4IY40 B4QQX6 A0A195AZF0 A0A1J1ISL4 A0A195EE01 A0A195FYC2 A0A2J7PUR1 A0A1B6GS71 A0A0A9VWL5 J3JVK8 U4U9L0 A0A0K8SZS3 A0A195D5T8 A0A0T6B504 A0A026WT83 A0A0E3M394 A0A3L8DG03 A0A0C9RCT3 A0A1Y1MEQ4 A0A2S2QCD2 A0A2Z4N531 A0A232ESB3 J9JNW2
PDB
5ZNT
E-value=0,
Score=2085
Ontologies
GO
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.989798
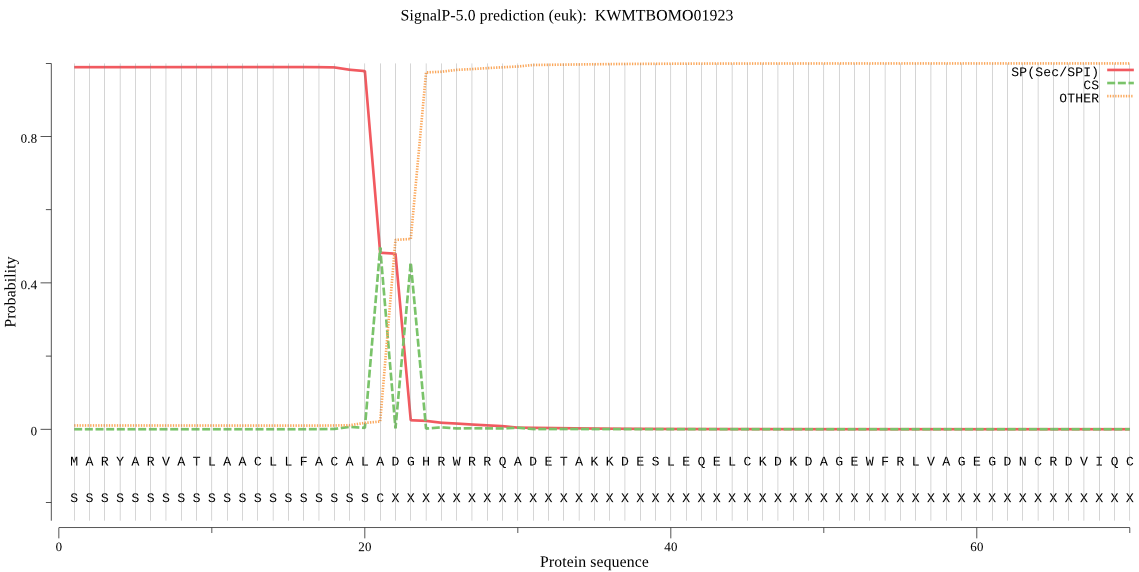
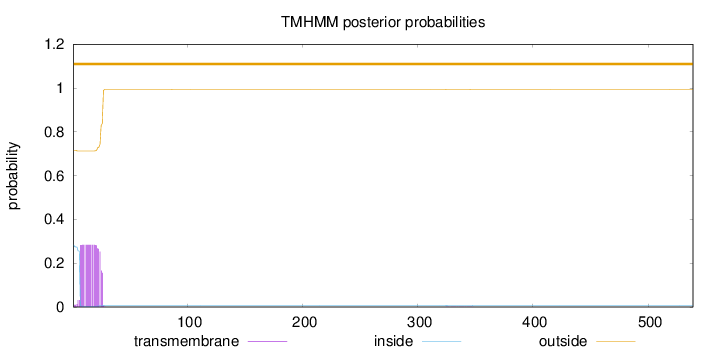
Length:
539
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.47405999999998
Exp number, first 60 AAs:
5.46424
Total prob of N-in:
0.28455
outside
1 - 539
Population Genetic Test Statistics
Pi
264.543574
Theta
193.836026
Tajima's D
1.090004
CLR
0.025068
CSRT
0.688165591720414
Interpretation
Uncertain
