Pre Gene Modal
BGIBMGA005934
Annotation
PREDICTED:_integrator_complex_subunit_2_[Amyelois_transitella]
Full name
Integrator complex subunit 2
Location in the cell
Cytoplasmic Reliability : 1.356 Extracellular Reliability : 1.381
Sequence
CDS
ATGGATATTGAATTCATGAAACCAGTTCAACCACACGTTTTCAAAGCCCTAAAAGATGTTGATATTCCGGGTCTTATGAAATGTAGTCCTGATGAAATAAGACCCATAATACCATGTCTTGTCCGAATGGCCTTGATATCTCCATTAGATATCACCAGATACTGTGCAGAAGCTAAGAAAGATATACTTACACTACTTTCAGGCATTGATTTAGTAAATTTCATAGTTTCACTTCTATCTATAGAATTTCATGTTCTAGAAGCAGATGTAAAAAAAGAACAACAAATGAGGCAAAAAAGTGGATCGCAATGCATTGAGTCAACACTAATTCCAAATATAATTAATGGTATTGCAAGTGATTTTGAACAGTCTGATTCTTCTAGGAGGGTTCGCCTTGTTCTATCAGAACTATTACAAATGCAAGCACAATTAGTTGAGTTCAATCAGACAAAAAATACAAATGCGGATTATACAATTAAGCCCTCTGAATTATTTGATAATGATGTGTATTTAGAAGAAATCACAGATGTTATATGTATAAGTTTAGCTGAAATACCGAATTTACTAAATATCTGTGATGTGGTTGAGGTTCTCCTTTATGTAAATAAGGGCCCTTCTATAATATCATGGGTGGTAGCAAATCAACCTGACATGTTGCAGGAAGTAGCGGAATCACTTGTATTAAATGCTGAACGAAGTGAAGAAGGTGGCATCAGAGCAAAGGCATTAGCTACCTTATGTGCTGTATTTCCACCTATTGCTGCCACCATAAGAGCAAAAGCAGCATCTGCTTCACGTCTACCATCTTTAGTCATTAATTTAACCTTGACTCACCATGAAGATGATCTTGTGTCATTTATATCAGGTCTGTTACTTGGATCTGACCAAACTACTAGAGGATGGTTTGCTACATTTTTACGAAATTCACACAAAAGAGGTAAAGGTGATGGTCATGCTGCATTGACAAAGTTAAGACAAGAATTATTAATTCGTCTAAAAGCTGCCACAGCTGGAGTAGATGCTTCAGCTTTACTAAGATTGTACTGTGCATTGAGGGGCATTGCCAGTATTAAGTTTCAGGATGATGAAGTAGCTTGGTTACTAAGACTAGTGACACAGAAACCTCCCCCAACCCCTGCTGGAGTCAGATTTGTCTCATTAAGTCTATGTATGATTTTAGCATGTCCATCCTTAATGGCTGCTCCAGATCATGAGAAAAAAGCTATAGAATGGGTCCAATGGTTGGTAAAAGAGGAAGCCTACTTCGAAAGCACTTCTGGTGTTACTGCATCTTTTGGTGAGATGTTGTTGCTGATTGCCATACATTTCCATTCCGGACAACTATCGGCAGTAGGTGAACTAGTGTGTGCAACACTTGGTATGCGTGTTCCAGTAAGACCTAATGGGCTGGCTAGAATAAAACAAGCCTTTACACAAGAAATATTTACAGAGCAGGTTGTGACAGCTCATGCAGTCAAAGTTCCTGTAACAGCTAATCTCAATAGCAATATTCCAGGATATTTGCCAGTTCATTGTATCCATCAACTACTAAAATCTAGAGCATTTTCGAAACACAAAGTGCCAATTAAAAATTGGATTTACCGACAAATATGTAACTGTACAGCACCTCTACATCCTGTTATGCCAGCACTGGTGGAGGTGTATGTCAATTCTATCCTTGTTGTAAACAACAAGGGTACTAACGAATATGTTAACAAACCTATAATGGAAGAAGAAATCCGTAAGGTTTTTAGAAACTCTATTTTTGGAGCAAATTTTGATGTAAAAAATAAATCAATAACTCCCATGGAAATAGATAATACTGAACAATCTGTAGAAATAAGACTAGAAAAACCAACGTTAGCTTCGCAGTTATTATTGATTTATTATTTGCTACTCTACGAAGATGTGCGATTGTCAAACACGGCAACGTTGCTTGCTAACGGTAGGAAGGTGAAAAGCTATTCAGCAGCATTTTTATCAGAATTGCCAATAAAATATCTATTGCACCAAGCCCAAAAAAATCAACTCAGTTATGGAGGTCTATTTAGTCCACTATTACGACTATTAGCGACACACTTCCCTCAATTATCTTTAGTCGATGACTGGATGGACGACCAAGTGTTTGCTGATATGAGTCGCTGTTATGTCGATTTCAATCTTTCCGAAACCACAATAAACGATGGATTCCTAAACATTGATGATAATCCGTATAAAACAGGAAAAGTTTTAAAAGCGATGTTAAGTAAAAATCCTACAGACATTTGGCCTTTTGCTGAGATATTCGTTAAATATATAAAAAGTGTATTAGGTGATAAAGTTCCTAGACATATACAAGAGTTATATCGGGAAGTTTGGTTACGACTCAATACTGTTTTACCCCGATGTTTGTGGATTATGACGATCAACGCTCTTCTAGATATTAACGGCATGGGTATTAAGAATGTTACTATTACACAAGAAAATGTTTTAGTTGATCCTTTACAAGTGCTGCGTTGCGATATGCGAGTATACAGATGTGGACCTATATTAAAAATTATTTTACGAATTCTCGAAGCAAGTTTGGCAGCTTCTAGAAGTCAACTATCACGTCACTTGCTCGATAAACCTTTATTAGAAAAAAGTGGCCAACTAACTTCCGATTCAGAAAGAGAAGAACTAAAAAATGCTTTAGTTGCAGCTCAAGAGAGCGCGGCCCTTCAAATATTATTAGAAGCCTGTCTAGAAACACCAGATGACCAAAAAAAACCTGAACTTATGTGGTCCCTTCGCGAAGTCCGAAGCATAATTTGTTCTTTTTTACATCAAATATTTATATCAGAACCATCTCTAGCAAAACTTGTGCATTTTCAAGGTTATCCCCGAGAACTTTTACCAGTAACAGTACAAGGCATTCCATCAATGCACATCTGTTTAGACTTTATACCAGAGCTATTAAGCCAAGCCTCTCTTGAAAAGCAAATATTTGCTGTTGACCTAGTATCTCATTTGTCCATCCAATATGCTTTACCTAAAGCAATGTCAATAGCCAGACTATGTGTGAATACTCTTTCAACACTATTATCTGTATTACCTAGTGATTTGCGTCTGGAATTATTCCAACCGGTTCTCAAGTCTCTGGAAAGGATATGCATCGCCTTTCCATCACTACTAGAAGATATTACATCTTTACTATTGCAGCTTGGCAGGATATGCGAATCCCAAGCGTCACTGGGACACTGTTGGAATGACACTCCCATATTAGGCGAAACTGCCTACGTTGATTCTGAAAGGCAACCTGAAACTAAAGTTCTAGTAGCGGAAGTCCTCAGTAGAGATATAAGGGTAACAATGAATGAAATTGTAAAAAAAGCTGTATTAAATGATAAATTATATTAA
Protein
MDIEFMKPVQPHVFKALKDVDIPGLMKCSPDEIRPIIPCLVRMALISPLDITRYCAEAKKDILTLLSGIDLVNFIVSLLSIEFHVLEADVKKEQQMRQKSGSQCIESTLIPNIINGIASDFEQSDSSRRVRLVLSELLQMQAQLVEFNQTKNTNADYTIKPSELFDNDVYLEEITDVICISLAEIPNLLNICDVVEVLLYVNKGPSIISWVVANQPDMLQEVAESLVLNAERSEEGGIRAKALATLCAVFPPIAATIRAKAASASRLPSLVINLTLTHHEDDLVSFISGLLLGSDQTTRGWFATFLRNSHKRGKGDGHAALTKLRQELLIRLKAATAGVDASALLRLYCALRGIASIKFQDDEVAWLLRLVTQKPPPTPAGVRFVSLSLCMILACPSLMAAPDHEKKAIEWVQWLVKEEAYFESTSGVTASFGEMLLLIAIHFHSGQLSAVGELVCATLGMRVPVRPNGLARIKQAFTQEIFTEQVVTAHAVKVPVTANLNSNIPGYLPVHCIHQLLKSRAFSKHKVPIKNWIYRQICNCTAPLHPVMPALVEVYVNSILVVNNKGTNEYVNKPIMEEEIRKVFRNSIFGANFDVKNKSITPMEIDNTEQSVEIRLEKPTLASQLLLIYYLLLYEDVRLSNTATLLANGRKVKSYSAAFLSELPIKYLLHQAQKNQLSYGGLFSPLLRLLATHFPQLSLVDDWMDDQVFADMSRCYVDFNLSETTINDGFLNIDDNPYKTGKVLKAMLSKNPTDIWPFAEIFVKYIKSVLGDKVPRHIQELYREVWLRLNTVLPRCLWIMTINALLDINGMGIKNVTITQENVLVDPLQVLRCDMRVYRCGPILKIILRILEASLAASRSQLSRHLLDKPLLEKSGQLTSDSEREELKNALVAAQESAALQILLEACLETPDDQKKPELMWSLREVRSIICSFLHQIFISEPSLAKLVHFQGYPRELLPVTVQGIPSMHICLDFIPELLSQASLEKQIFAVDLVSHLSIQYALPKAMSIARLCVNTLSTLLSVLPSDLRLELFQPVLKSLERICIAFPSLLEDITSLLLQLGRICESQASLGHCWNDTPILGETAYVDSERQPETKVLVAEVLSRDIRVTMNEIVKKAVLNDKLY
Summary
Description
Component of the Integrator (INT) complex, a complex involved in the small nuclear RNAs (snRNA) U1 and U2 transcription and in their 3'-box-dependent processing. The Integrator complex is associated with the C-terminal domain (CTD) of RNA polymerase II largest subunit (POLR2A) and is recruited to the U1 and U2 snRNAs genes (Probable). Mediates recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (PubMed:23904267).
Subunit
Belongs to the multiprotein complex Integrator, at least composed of INTS1, INTS2, INTS3, INTS4, INTS5, INTS6, INTS7, INTS8, INTS9/RC74, INTS10, INTS11/CPSF3L and INTS12.
Similarity
Belongs to the Integrator subunit 2 family.
Keywords
Complete proteome
Cytoplasm
Membrane
Nucleus
Polymorphism
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Integrator complex subunit 2
sequence variant In dbSNP:rs606072.
sequence variant In dbSNP:rs606072.
Uniprot
H9J8U2
A0A2W1BYU7
A0A2A4K2P8
A0A2H1VHU9
A0A194RCI7
A0A194PKL3
+ More
A0A212ETV4 A0A0L7LLW8 E2BUF7 F4W7M7 A0A151WJ29 A0A2J7Q795 A0A0C9Q7Q4 A0A3L8D732 A0A158NPM6 A0A067QT10 E9I8Q0 E2AXQ5 A0A195ESH2 A0A2A3E5M2 A0A195ATS2 A0A026W1K7 A0A088AVZ3 A0A195EM91 A0A0L7QP88 A0A154PLI3 A0A0J7KRV1 A0A151IMN2 K7J951 A0A232F4L6 A0A310SP61 U5ETJ7 E0VMM5 A0A1Q3F509 A0A0P6IWG5 B0X5M1 A0A1S4FSB1 A0A182JDV3 Q16QV6 A0A1W4X6V2 A0A182VEN4 A0A182LFR1 Q5TU39 A0A182TYT0 A0A182N622 A0A182JZH3 A0A182W082 A0A182HU99 A0A182WRH8 A0A084VRE5 A0A182MDC8 A0A1B6CUF6 A0A182Q203 A0A182Y4B8 A0A182RNP3 A0A182G3Z7 A0A1Q3F584 A0A182PAA8 A0A0A9VRS4 A0A1Y1MAS2 A0A023F3Y1 D2A4V3 A0A336MQH9 A0A336LMB4 A0A1J1ID50 A0A1S3D595 A0A0M9A8K3 T1IXD4 E9G4S7 A0A087TK98 A0A0P5A9S8 A0A0P5K1P9 A0A0P5MQR3 A0A182SC39 A0A0P6IGV9 A0A0P6CPE4 A0A131XRZ2 A0A0N8CGX1 A0A147BNQ3 A0A2R5L534 A0A0P5VN97 A0A162N9D3 A0A1Z5KUH2 A0A0T6B323 A0A0P4Y623 A0A1Z5L4M5 A0A2L2YLM1 A0A2K5Y2N4 A0A2K6NSA0 F6U3W7 A0A024QZ48 A0A341CFJ0 A0A2U4C6E2 A0A340Y0F6 A0A2Y9ETB7 A0A2J8W7E3 J3KMZ7 Q9H0H0 A0A2Y9QYN7
A0A212ETV4 A0A0L7LLW8 E2BUF7 F4W7M7 A0A151WJ29 A0A2J7Q795 A0A0C9Q7Q4 A0A3L8D732 A0A158NPM6 A0A067QT10 E9I8Q0 E2AXQ5 A0A195ESH2 A0A2A3E5M2 A0A195ATS2 A0A026W1K7 A0A088AVZ3 A0A195EM91 A0A0L7QP88 A0A154PLI3 A0A0J7KRV1 A0A151IMN2 K7J951 A0A232F4L6 A0A310SP61 U5ETJ7 E0VMM5 A0A1Q3F509 A0A0P6IWG5 B0X5M1 A0A1S4FSB1 A0A182JDV3 Q16QV6 A0A1W4X6V2 A0A182VEN4 A0A182LFR1 Q5TU39 A0A182TYT0 A0A182N622 A0A182JZH3 A0A182W082 A0A182HU99 A0A182WRH8 A0A084VRE5 A0A182MDC8 A0A1B6CUF6 A0A182Q203 A0A182Y4B8 A0A182RNP3 A0A182G3Z7 A0A1Q3F584 A0A182PAA8 A0A0A9VRS4 A0A1Y1MAS2 A0A023F3Y1 D2A4V3 A0A336MQH9 A0A336LMB4 A0A1J1ID50 A0A1S3D595 A0A0M9A8K3 T1IXD4 E9G4S7 A0A087TK98 A0A0P5A9S8 A0A0P5K1P9 A0A0P5MQR3 A0A182SC39 A0A0P6IGV9 A0A0P6CPE4 A0A131XRZ2 A0A0N8CGX1 A0A147BNQ3 A0A2R5L534 A0A0P5VN97 A0A162N9D3 A0A1Z5KUH2 A0A0T6B323 A0A0P4Y623 A0A1Z5L4M5 A0A2L2YLM1 A0A2K5Y2N4 A0A2K6NSA0 F6U3W7 A0A024QZ48 A0A341CFJ0 A0A2U4C6E2 A0A340Y0F6 A0A2Y9ETB7 A0A2J8W7E3 J3KMZ7 Q9H0H0 A0A2Y9QYN7
Pubmed
19121390
28756777
26354079
22118469
26227816
20798317
+ More
21719571 30249741 21347285 24845553 21282665 24508170 20075255 28648823 20566863 26999592 17510324 20966253 12364791 14747013 17210077 24438588 25244985 26483478 25401762 28004739 25474469 18362917 19820115 21292972 29652888 28528879 26561354 25362486 19892987 11181995 16625196 10574462 11230166 16239144 23904267
21719571 30249741 21347285 24845553 21282665 24508170 20075255 28648823 20566863 26999592 17510324 20966253 12364791 14747013 17210077 24438588 25244985 26483478 25401762 28004739 25474469 18362917 19820115 21292972 29652888 28528879 26561354 25362486 19892987 11181995 16625196 10574462 11230166 16239144 23904267
EMBL
BABH01005051
BABH01005052
BABH01005053
KZ149895
PZC78825.1
NWSH01000193
+ More
PCG78525.1 ODYU01002642 SOQ40417.1 KQ460398 KPJ15184.1 KQ459601 KPI93867.1 AGBW02012530 OWR44923.1 JTDY01000698 KOB76181.1 GL450640 EFN80684.1 GL887844 EGI69900.1 KQ983049 KYQ47852.1 NEVH01017447 PNF24457.1 GBYB01010193 JAG79960.1 QOIP01000012 RLU16031.1 ADTU01022615 KK853054 KDR12003.1 GL761642 EFZ23053.1 GL443628 EFN61801.1 KQ981986 KYN31195.1 KZ288360 PBC27037.1 KQ976745 KYM75384.1 KK107488 EZA49947.1 KQ978730 KYN28994.1 KQ414843 KOC60311.1 KQ434971 KZC12729.1 LBMM01003940 KMQ92974.1 KQ977026 KYN06219.1 NNAY01001028 OXU25429.1 KQ761546 OAD57408.1 GANO01001887 JAB57984.1 DS235317 EEB14631.1 GFDL01012394 JAV22651.1 GDUN01000479 JAN95440.1 DS232386 EDS40958.1 CH477729 EAT36795.1 AAAB01008859 EAL40780.3 APCN01000613 ATLV01015610 KE525023 KFB40539.1 AXCM01000851 GEDC01020244 JAS17054.1 AXCN02000193 JXUM01041643 KQ561292 KXJ79094.1 GFDL01012329 JAV22716.1 GBHO01044527 JAF99076.1 GEZM01036155 JAV82932.1 GBBI01003053 JAC15659.1 KQ971344 EFA05171.1 UFQS01001992 UFQT01001992 SSX12898.1 SSX32340.1 UFQT01000056 SSX19126.1 CVRI01000047 CRK98160.1 KQ435711 KOX79427.1 JH431646 GL732532 EFX85507.1 KK115611 KFM65537.1 GDIP01214515 JAJ08887.1 GDIQ01190494 GDIQ01190493 GDIQ01190492 GDIQ01190491 GDIQ01190490 JAK61233.1 GDIQ01164159 JAK87566.1 GDIQ01004719 JAN90018.1 GDIP01011465 JAM92250.1 GEFM01006881 JAP68915.1 GDIP01132879 JAL70835.1 GEGO01002997 JAR92407.1 GGLE01000498 MBY04624.1 GDIP01097726 JAM05989.1 LRGB01000568 KZS17794.1 GFJQ02008214 JAV98755.1 LJIG01016082 KRT81668.1 GDIP01233063 JAI90338.1 GFJQ02004604 JAW02366.1 IAAA01035255 LAA08996.1 CH471179 EAW51436.1 NDHI03003397 PNJ65694.1 PNJ65695.1 AC060798 KF456301 AB033113 AL136800 CR533582 BK005721
PCG78525.1 ODYU01002642 SOQ40417.1 KQ460398 KPJ15184.1 KQ459601 KPI93867.1 AGBW02012530 OWR44923.1 JTDY01000698 KOB76181.1 GL450640 EFN80684.1 GL887844 EGI69900.1 KQ983049 KYQ47852.1 NEVH01017447 PNF24457.1 GBYB01010193 JAG79960.1 QOIP01000012 RLU16031.1 ADTU01022615 KK853054 KDR12003.1 GL761642 EFZ23053.1 GL443628 EFN61801.1 KQ981986 KYN31195.1 KZ288360 PBC27037.1 KQ976745 KYM75384.1 KK107488 EZA49947.1 KQ978730 KYN28994.1 KQ414843 KOC60311.1 KQ434971 KZC12729.1 LBMM01003940 KMQ92974.1 KQ977026 KYN06219.1 NNAY01001028 OXU25429.1 KQ761546 OAD57408.1 GANO01001887 JAB57984.1 DS235317 EEB14631.1 GFDL01012394 JAV22651.1 GDUN01000479 JAN95440.1 DS232386 EDS40958.1 CH477729 EAT36795.1 AAAB01008859 EAL40780.3 APCN01000613 ATLV01015610 KE525023 KFB40539.1 AXCM01000851 GEDC01020244 JAS17054.1 AXCN02000193 JXUM01041643 KQ561292 KXJ79094.1 GFDL01012329 JAV22716.1 GBHO01044527 JAF99076.1 GEZM01036155 JAV82932.1 GBBI01003053 JAC15659.1 KQ971344 EFA05171.1 UFQS01001992 UFQT01001992 SSX12898.1 SSX32340.1 UFQT01000056 SSX19126.1 CVRI01000047 CRK98160.1 KQ435711 KOX79427.1 JH431646 GL732532 EFX85507.1 KK115611 KFM65537.1 GDIP01214515 JAJ08887.1 GDIQ01190494 GDIQ01190493 GDIQ01190492 GDIQ01190491 GDIQ01190490 JAK61233.1 GDIQ01164159 JAK87566.1 GDIQ01004719 JAN90018.1 GDIP01011465 JAM92250.1 GEFM01006881 JAP68915.1 GDIP01132879 JAL70835.1 GEGO01002997 JAR92407.1 GGLE01000498 MBY04624.1 GDIP01097726 JAM05989.1 LRGB01000568 KZS17794.1 GFJQ02008214 JAV98755.1 LJIG01016082 KRT81668.1 GDIP01233063 JAI90338.1 GFJQ02004604 JAW02366.1 IAAA01035255 LAA08996.1 CH471179 EAW51436.1 NDHI03003397 PNJ65694.1 PNJ65695.1 AC060798 KF456301 AB033113 AL136800 CR533582 BK005721
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000008237 UP000007755 UP000075809 UP000235965 UP000279307 UP000005205 UP000027135 UP000000311 UP000078541 UP000242457 UP000078540 UP000053097 UP000005203 UP000078492 UP000053825 UP000076502 UP000036403 UP000078542 UP000002358 UP000215335 UP000009046 UP000002320 UP000075880 UP000008820 UP000192223 UP000075903 UP000075882 UP000007062 UP000075902 UP000075884 UP000075881 UP000075920 UP000075840 UP000076407 UP000030765 UP000075883 UP000075886 UP000076408 UP000075900 UP000069940 UP000249989 UP000075885 UP000007266 UP000183832 UP000079169 UP000053105 UP000000305 UP000054359 UP000075901 UP000076858 UP000233140 UP000233200 UP000002281 UP000252040 UP000245320 UP000265300 UP000248484 UP000005640 UP000248480
UP000008237 UP000007755 UP000075809 UP000235965 UP000279307 UP000005205 UP000027135 UP000000311 UP000078541 UP000242457 UP000078540 UP000053097 UP000005203 UP000078492 UP000053825 UP000076502 UP000036403 UP000078542 UP000002358 UP000215335 UP000009046 UP000002320 UP000075880 UP000008820 UP000192223 UP000075903 UP000075882 UP000007062 UP000075902 UP000075884 UP000075881 UP000075920 UP000075840 UP000076407 UP000030765 UP000075883 UP000075886 UP000076408 UP000075900 UP000069940 UP000249989 UP000075885 UP000007266 UP000183832 UP000079169 UP000053105 UP000000305 UP000054359 UP000075901 UP000076858 UP000233140 UP000233200 UP000002281 UP000252040 UP000245320 UP000265300 UP000248484 UP000005640 UP000248480
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
H9J8U2
A0A2W1BYU7
A0A2A4K2P8
A0A2H1VHU9
A0A194RCI7
A0A194PKL3
+ More
A0A212ETV4 A0A0L7LLW8 E2BUF7 F4W7M7 A0A151WJ29 A0A2J7Q795 A0A0C9Q7Q4 A0A3L8D732 A0A158NPM6 A0A067QT10 E9I8Q0 E2AXQ5 A0A195ESH2 A0A2A3E5M2 A0A195ATS2 A0A026W1K7 A0A088AVZ3 A0A195EM91 A0A0L7QP88 A0A154PLI3 A0A0J7KRV1 A0A151IMN2 K7J951 A0A232F4L6 A0A310SP61 U5ETJ7 E0VMM5 A0A1Q3F509 A0A0P6IWG5 B0X5M1 A0A1S4FSB1 A0A182JDV3 Q16QV6 A0A1W4X6V2 A0A182VEN4 A0A182LFR1 Q5TU39 A0A182TYT0 A0A182N622 A0A182JZH3 A0A182W082 A0A182HU99 A0A182WRH8 A0A084VRE5 A0A182MDC8 A0A1B6CUF6 A0A182Q203 A0A182Y4B8 A0A182RNP3 A0A182G3Z7 A0A1Q3F584 A0A182PAA8 A0A0A9VRS4 A0A1Y1MAS2 A0A023F3Y1 D2A4V3 A0A336MQH9 A0A336LMB4 A0A1J1ID50 A0A1S3D595 A0A0M9A8K3 T1IXD4 E9G4S7 A0A087TK98 A0A0P5A9S8 A0A0P5K1P9 A0A0P5MQR3 A0A182SC39 A0A0P6IGV9 A0A0P6CPE4 A0A131XRZ2 A0A0N8CGX1 A0A147BNQ3 A0A2R5L534 A0A0P5VN97 A0A162N9D3 A0A1Z5KUH2 A0A0T6B323 A0A0P4Y623 A0A1Z5L4M5 A0A2L2YLM1 A0A2K5Y2N4 A0A2K6NSA0 F6U3W7 A0A024QZ48 A0A341CFJ0 A0A2U4C6E2 A0A340Y0F6 A0A2Y9ETB7 A0A2J8W7E3 J3KMZ7 Q9H0H0 A0A2Y9QYN7
A0A212ETV4 A0A0L7LLW8 E2BUF7 F4W7M7 A0A151WJ29 A0A2J7Q795 A0A0C9Q7Q4 A0A3L8D732 A0A158NPM6 A0A067QT10 E9I8Q0 E2AXQ5 A0A195ESH2 A0A2A3E5M2 A0A195ATS2 A0A026W1K7 A0A088AVZ3 A0A195EM91 A0A0L7QP88 A0A154PLI3 A0A0J7KRV1 A0A151IMN2 K7J951 A0A232F4L6 A0A310SP61 U5ETJ7 E0VMM5 A0A1Q3F509 A0A0P6IWG5 B0X5M1 A0A1S4FSB1 A0A182JDV3 Q16QV6 A0A1W4X6V2 A0A182VEN4 A0A182LFR1 Q5TU39 A0A182TYT0 A0A182N622 A0A182JZH3 A0A182W082 A0A182HU99 A0A182WRH8 A0A084VRE5 A0A182MDC8 A0A1B6CUF6 A0A182Q203 A0A182Y4B8 A0A182RNP3 A0A182G3Z7 A0A1Q3F584 A0A182PAA8 A0A0A9VRS4 A0A1Y1MAS2 A0A023F3Y1 D2A4V3 A0A336MQH9 A0A336LMB4 A0A1J1ID50 A0A1S3D595 A0A0M9A8K3 T1IXD4 E9G4S7 A0A087TK98 A0A0P5A9S8 A0A0P5K1P9 A0A0P5MQR3 A0A182SC39 A0A0P6IGV9 A0A0P6CPE4 A0A131XRZ2 A0A0N8CGX1 A0A147BNQ3 A0A2R5L534 A0A0P5VN97 A0A162N9D3 A0A1Z5KUH2 A0A0T6B323 A0A0P4Y623 A0A1Z5L4M5 A0A2L2YLM1 A0A2K5Y2N4 A0A2K6NSA0 F6U3W7 A0A024QZ48 A0A341CFJ0 A0A2U4C6E2 A0A340Y0F6 A0A2Y9ETB7 A0A2J8W7E3 J3KMZ7 Q9H0H0 A0A2Y9QYN7
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus membrane
Cytoplasm
Cytoplasm
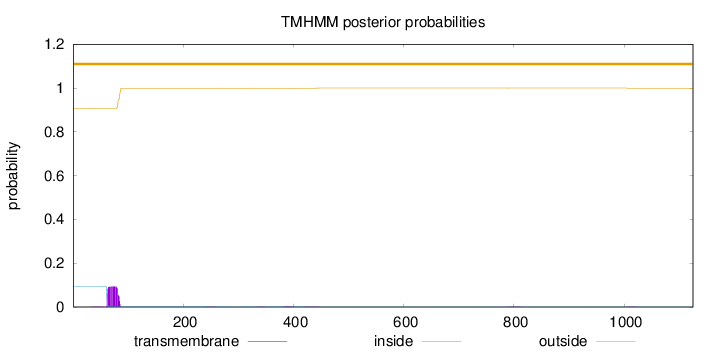
Length:
1125
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.07223000000003
Exp number, first 60 AAs:
0.0064
Total prob of N-in:
0.09286
outside
1 - 1125
Population Genetic Test Statistics
Pi
158.87166
Theta
195.697292
Tajima's D
0.327477
CLR
0.207023
CSRT
0.459977001149942
Interpretation
Uncertain
