Gene
KWMTBOMO01919
Pre Gene Modal
BGIBMGA005935
Annotation
PREDICTED:_frizzled-7-B_[Amyelois_transitella]
Full name
Frizzled-7-B
+ More
Frizzled-7-A
Frizzled-7
Frizzled-7-A
Frizzled-7
Location in the cell
Cytoplasmic Reliability : 1.151 Nuclear Reliability : 2.775
Sequence
CDS
ATGGCAAAATTTTGGAAACATTATGTTATTATATTCAGTCTGTTATGTCTTTTTGCGTTAACAAAAGGTTCTCGAGAGATTGTTTATCAAGGCGATAGTCTACCTCAACATGGACGTTGTGAACCCATTACCATTCAGTTTTGTCAGAAATTGCGTTATAATCAAACCATTTTTCCGAATATTTTAAATCAAGCACGACAAGAAGATGCAGCCGCAAACATGCTTCTGTTTACAACTCTCATTAAACTGAATTGCTCTCCTGATCTAAGATTCTTCTTGTGTTCAGTTTATGCACCCGTTTGCACGATACTAGATTCTGCGATACCGCCTTGTCGTCATTTATGTGAAGCTGCAAAACAAAGCTGTGATGTCGTCATACGAAAATTTGATTTTCCTTGGCCCCCTGAGCTTGAATGTTCTGAATTTCCTGAAGTATCTGATAATAACATATGTGTTGGTGACAATAATACTGCCCATGAATCTCGAAAACGCACTGATACCAAGTTTCGTAAGGTTGACTTCAAATCTAACATGGAGAGAGATCCGACTAGATTGCATGAGTTTGTGTGCCCAGTGCAGTTTAAAGTACCAAGACATTTAGATCTTGAATACTCTTTTAAAGTTGGAAATAGAGTTGAACATGACTGTGGAGCCCCATGCAATGGAATGTTCTTCAGTAAAGATGAAAAGAATTTTTCAAGGCTCTGGATTGGAATCTGGGCAGTCCTGTGTGCACTCAGTTGCCTTTTTACTGTGCTAACATTTTTAATAGATACAGATAGATTCAGGTATCCAGAAAGACCCATAATATTTTTGTCAGTTTGTTATCTAATGGTAGCTATTGCCTACATAATGGGTTGGAGTGCAGGTGATAATATAAGCTGCCAAGGGCCTTTTGTTTCTGCTGTCAGTGGAACAAGATTACCCAACATATCAGTTATCACACAAGGTACAAAACATGAGCCATGCACAATACTGTTTATGATAGTATATTTCTTTAGTATGGCTTCGAGTATATGGTGGGTCATATTAACCCTGACATGGTTTTTGGCTGCTGGCCTAAAATGGGGACATGAAGCAATAGAAGCAAATTCCCAATATTTTCATCTTGCTGCTTGGGCAGTACCAGCTGTGAAGACAATATCTATTCTAGCAATGGGGAAAGTAGATGGTGATGTACTGTCAGGTGTATGTTACGTTGGATTATGGGATGCAGAAGCTTTAAGAGGATTTGTGTTGGCACCTTTATGTGTCTATTTAGTGTTTGGAACAATCTTTCTTCTAGCTGGCTTTGTCTCACTGTTTCGGATCAGAACTGTCATGAAACATGATGGGACTAAAACTGATAAATTAGAGAAACTGATGATTCGCATTGGTGTGTTTGGTGTACTATACACCGTGCCTGCCCTCATAGTCATAGCATGTTTGTTCTATGAACAAGTTAATTTTGACAAATGGATGATTGCATGGCAACGAGACATGTGTTCGATACGACAATACTCAATTCCTTGTCCGATAATTCATGAAGAGATAGAAGGACCAAAGTTTGAAATGTTTATGATTAAATATCTAATGACAATGATTGTTGGTATAACATCTAGCTTTTGGATATGGTCTGGTAAAACTTTGATATCATGGTCTCAATTTTTTGACAAAGTTAAAGGTCGCAGAGTAGAAGCCTATGTGTAA
Protein
MAKFWKHYVIIFSLLCLFALTKGSREIVYQGDSLPQHGRCEPITIQFCQKLRYNQTIFPNILNQARQEDAAANMLLFTTLIKLNCSPDLRFFLCSVYAPVCTILDSAIPPCRHLCEAAKQSCDVVIRKFDFPWPPELECSEFPEVSDNNICVGDNNTAHESRKRTDTKFRKVDFKSNMERDPTRLHEFVCPVQFKVPRHLDLEYSFKVGNRVEHDCGAPCNGMFFSKDEKNFSRLWIGIWAVLCALSCLFTVLTFLIDTDRFRYPERPIIFLSVCYLMVAIAYIMGWSAGDNISCQGPFVSAVSGTRLPNISVITQGTKHEPCTILFMIVYFFSMASSIWWVILTLTWFLAAGLKWGHEAIEANSQYFHLAAWAVPAVKTISILAMGKVDGDVLSGVCYVGLWDAEALRGFVLAPLCVYLVFGTIFLLAGFVSLFRIRTVMKHDGTKTDKLEKLMIRIGVFGVLYTVPALIVIACLFYEQVNFDKWMIAWQRDMCSIRQYSIPCPIIHEEIEGPKFEMFMIKYLMTMIVGITSSFWIWSGKTLISWSQFFDKVKGRRVEAYV
Summary
Description
Receptor for Wnt proteins. Acts in both canonical and non-canonical Wnt pathways. Although different papers report differing Wnt preferences, wnt5a, wnt8b and wnt11 have been proposed as synergists. In the canonical Wnt pathway, acts via beta-catenin to promote the expression of the dorsal genes siamois, twin and nodal3 and to establish the dorsal axis of the embryo and induce dorsal mesoderm formation. In a non-canonical Wnt/planar cell polarity (PCP) pathway, acts with sdc4 and dvl2/dsh to regulate convergent extension cell movements during gastrulation. Triggers phosphorylation of dvl2/dsh and its translocation to the plasma membrane. In a third branch of Wnt signaling, acts in a non-canonical pathway via trimeric G proteins, and independently of dvl2/dsh, to recruit protein kinase C (PKC) to the membrane and thus activate PKC. PKC signaling controls cell sorting and tissue separation during gastrulation.
Receptor for Wnt proteins. Acts in both canonical and non-canonical Wnt pathways. Although different papers report differing Wnt preferences, wnt5a, wnt8b and wnt11 have been proposed as synergists. In the canonical Wnt pathway, acts via beta-catenin to promote the expression of the dorsal genes siamois, twin and nodal3 and to establish the dorsal axis of the embryo and induce dorsal mesoderm formation. In a non-canonical Wnt/planar cell polarity (PCP) pathway, acts with sdc4 and dvl2/dsh to regulate convergent extension movements in gastrulation. Triggers phosphorylation of dvl2/dsh and its translocation to the plasma membrane. In a third branch of Wnt signaling, acts in a non-canonical pathway via trimeric G proteins, and independently of dvl2/dsh, to recruit protein kinase C (PKC) to the membrane and thus activate PKC. PKC signaling controls cell sorting and tissue separation during gastrulation (By similarity).
Receptor for Wnt proteins. Acts in both canonical and non-canonical Wnt pathways. Although different papers report differing Wnt preferences, wnt5a, wnt8b and wnt11 have been proposed as synergists. In the canonical Wnt pathway, acts via beta-catenin to promote the expression of the dorsal genes siamois, twin and nodal3 and to establish the dorsal axis of the embryo and induce dorsal mesoderm formation. In a non-canonical Wnt/planar cell polarity (PCP) pathway, acts with sdc4 and dvl2/dsh to regulate convergent extension movements in gastrulation. Triggers phosphorylation of dvl2/dsh and its translocation to the plasma membrane. In a third branch of Wnt signaling, acts in a non-canonical pathway via trimeric G proteins, and independently of dvl2/dsh, to recruit protein kinase C (PKC) to the membrane and thus activate PKC. PKC signaling controls cell sorting and tissue separation during gastrulation (By similarity).
Subunit
Interacts with wnt11 and sdc4. The extracellular domain interacts with the extracellular domain of pcdh8/papc (By similarity). Interacts (via C-terminus) with dvl1 (via PDZ domain).
Interacts with wnt11 and sdc4. The extracellular domain interacts with the extracellular domain of pcdh8/papc.
Interacts with wnt11 and sdc4. The extracellular domain interacts with the extracellular domain of pcdh8/papc (By similarity).
Interacts with wnt11 and sdc4. The extracellular domain interacts with the extracellular domain of pcdh8/papc.
Interacts with wnt11 and sdc4. The extracellular domain interacts with the extracellular domain of pcdh8/papc (By similarity).
Similarity
Belongs to the G-protein coupled receptor Fz/Smo family.
Keywords
Cell membrane
Developmental protein
Disulfide bond
Endosome
G-protein coupled receptor
Glycoprotein
Membrane
Receptor
Signal
Transducer
Transmembrane
Transmembrane helix
Wnt signaling pathway
Complete proteome
Reference proteome
Feature
chain Frizzled-7-B
Uniprot
H9J8U3
A0A2W1BZZ7
A0A2A4K442
A0A2H1VHU6
A0A1E1WUB8
A0A194RGX7
+ More
A0A0L7LLG7 A0A1B6FSU1 A0A067QHH7 A0A1B6DUK5 A0A195FKC0 K7IXM9 E2B9L1 E2AHT4 A0A023F262 A0A2A3EBH9 A0A088A6V2 A0A3L8DES9 A0A154PH96 A0A026WRU5 A0A1Y1NNJ7 A0A0A9VZ38 A0A0K8S350 U4U4I1 N6U9T1 D6WJY3 A0A143VP60 T1JLV3 A0A224XM69 A0A087T8F5 V5HFW8 B7PFK7 L7M581 A0A1W4XSY0 A0A2T7Q003 A0A131YWN6 V5SQJ2 E9G2J9 W5UR01 A0A2D0S5B5 Q98SI2 Q7SZR7 Q6NV44 V4CHR5 A0A3P8YED9 H3AZ03 A0A2K9RCG5 Q90ZT3 I3KZU0 A0A3N0XUZ5 A0A0P7TZS4 A0A3B4CXR7 Q8QFM3 A0A1S3JF10 C3YEA5 Q8AVJ9 A0A3P8ZXJ8 A0A3N0Y6C7 A0A1A7XST0 W5NMT4 A0A2U9AZV8 H3B255 W5MX40 A0A1L8EX17 A0A1W4Y9D2 A0A3P9C180 A0A3P9D6V3 Q9PUK8 A0A3B4FUC1 A0A3B4GD67 Q7ZU08 W5LU60 A0A3P8NK42 G7N8N7 A0A1W5B4Z9 A0A3B3CTI6 A0A2K6B3T9 A0A2K5URA6 Q5BL72 A0A3N0Y5I0 A0A1W4ZY33 A0A3P8QM51 A0A1U7QEK4 A0A2K5JJR2 I3JSE2 G1K3F4 F1SI18 A0A340Y6D5 A0A2I2U7V2 M3Z5C2 A0A1L8EPZ4 A0A1S3MVZ0 A0A250Y2V1 A0A341CGM7 H0XWH9 A0A2Y9N1H2 A0A3B4ZM92 H2MKU6 A0A1S3MQT5
A0A0L7LLG7 A0A1B6FSU1 A0A067QHH7 A0A1B6DUK5 A0A195FKC0 K7IXM9 E2B9L1 E2AHT4 A0A023F262 A0A2A3EBH9 A0A088A6V2 A0A3L8DES9 A0A154PH96 A0A026WRU5 A0A1Y1NNJ7 A0A0A9VZ38 A0A0K8S350 U4U4I1 N6U9T1 D6WJY3 A0A143VP60 T1JLV3 A0A224XM69 A0A087T8F5 V5HFW8 B7PFK7 L7M581 A0A1W4XSY0 A0A2T7Q003 A0A131YWN6 V5SQJ2 E9G2J9 W5UR01 A0A2D0S5B5 Q98SI2 Q7SZR7 Q6NV44 V4CHR5 A0A3P8YED9 H3AZ03 A0A2K9RCG5 Q90ZT3 I3KZU0 A0A3N0XUZ5 A0A0P7TZS4 A0A3B4CXR7 Q8QFM3 A0A1S3JF10 C3YEA5 Q8AVJ9 A0A3P8ZXJ8 A0A3N0Y6C7 A0A1A7XST0 W5NMT4 A0A2U9AZV8 H3B255 W5MX40 A0A1L8EX17 A0A1W4Y9D2 A0A3P9C180 A0A3P9D6V3 Q9PUK8 A0A3B4FUC1 A0A3B4GD67 Q7ZU08 W5LU60 A0A3P8NK42 G7N8N7 A0A1W5B4Z9 A0A3B3CTI6 A0A2K6B3T9 A0A2K5URA6 Q5BL72 A0A3N0Y5I0 A0A1W4ZY33 A0A3P8QM51 A0A1U7QEK4 A0A2K5JJR2 I3JSE2 G1K3F4 F1SI18 A0A340Y6D5 A0A2I2U7V2 M3Z5C2 A0A1L8EPZ4 A0A1S3MVZ0 A0A250Y2V1 A0A341CGM7 H0XWH9 A0A2Y9N1H2 A0A3B4ZM92 H2MKU6 A0A1S3MQT5
Pubmed
19121390
28756777
26354079
26227816
24845553
20075255
+ More
20798317 25474469 30249741 24508170 28004739 25401762 26823975 23537049 18362917 19820115 25765539 25576852 26830274 24012522 21292972 23127152 11287199 23594743 23254933 25069045 9215903 29337984 25186727 12049772 12351181 18563158 10446283 10899005 11477687 14636582 27762356 10751186 10862746 10727861 10906785 10990458 11291855 11677610 15272309 16604063 25329095 22002653 29451363 20431018 30723633 17975172 28087693 17554307
20798317 25474469 30249741 24508170 28004739 25401762 26823975 23537049 18362917 19820115 25765539 25576852 26830274 24012522 21292972 23127152 11287199 23594743 23254933 25069045 9215903 29337984 25186727 12049772 12351181 18563158 10446283 10899005 11477687 14636582 27762356 10751186 10862746 10727861 10906785 10990458 11291855 11677610 15272309 16604063 25329095 22002653 29451363 20431018 30723633 17975172 28087693 17554307
EMBL
BABH01005049
KU291212
AMR93992.1
KZ149895
PZC78827.1
NWSH01000193
+ More
PCG78523.1 ODYU01002642 SOQ40415.1 GDQN01000490 JAT90564.1 KQ460398 KPJ15186.1 JTDY01000698 KOB76184.1 GECZ01030998 GECZ01016509 JAS38771.1 JAS53260.1 KK853378 KDR08023.1 GEDC01007942 JAS29356.1 KQ981490 KYN41115.1 AAZX01011865 GL446556 EFN87644.1 GL439579 EFN67013.1 GBBI01003040 JAC15672.1 KZ288312 PBC28411.1 QOIP01000009 RLU18831.1 KQ434893 KZC10688.1 KK107119 EZA58747.1 GEZM01001643 JAV97627.1 GBHO01044156 GBHO01036410 GBHO01010232 JAF99447.1 JAG07194.1 JAG33372.1 GBRD01018111 GDHC01020404 JAG47716.1 JAP98224.1 KB632006 ERL87962.1 APGK01034123 KB740904 ENN78450.1 KQ971342 EFA04653.1 LN849082 CRI73784.1 JH431585 GFTR01006734 JAW09692.1 KK113932 KFM61394.1 GANP01012075 JAB72393.1 ABJB010293041 DS703155 EEC05379.1 GACK01006785 JAA58249.1 PZQS01000001 PVD39003.1 GEDV01004944 JAP83613.1 KC690271 AHB53231.1 GL732530 EFX86266.1 JT416322 AHH42008.1 AJ301617 CAC37335.1 BC056280 AAH56280.2 CR407547 BC068322 AAH68322.2 KB200430 ESP01680.1 AFYH01133944 MG711509 AUT31435.1 AF336123 AAK49948.1 AERX01013317 RJVU01059775 ROJ66271.1 JARO02005224 KPP67133.1 BX927225 AF437316 AF336124 AF510101 AAL87635.1 AAM00192.1 AAN03844.1 GG666505 EEN61413.1 AF159106 BC042228 AAH42228.1 RJVU01051519 ROL41742.1 HADW01019494 SBP20894.1 AHAT01034647 CP026243 AWO97156.1 AFYH01065303 AHAT01004650 CM004482 OCT63845.1 AF039215 AJ243323 AF179213 AF114151 BC049397 AAH49397.1 CM001264 EHH21587.1 AQIA01015569 AC149036 CR759995 BC090579 AAH90579.1 AAZ06130.1 CAJ82817.1 RJVU01051648 ROL41476.1 AERX01048603 AAMC01057529 AEMK02000100 DQIR01145990 HDB01467.1 AANG04002752 AEYP01060373 CM004483 OCT61339.1 GFFV01001978 JAV37967.1 AAQR03032755
PCG78523.1 ODYU01002642 SOQ40415.1 GDQN01000490 JAT90564.1 KQ460398 KPJ15186.1 JTDY01000698 KOB76184.1 GECZ01030998 GECZ01016509 JAS38771.1 JAS53260.1 KK853378 KDR08023.1 GEDC01007942 JAS29356.1 KQ981490 KYN41115.1 AAZX01011865 GL446556 EFN87644.1 GL439579 EFN67013.1 GBBI01003040 JAC15672.1 KZ288312 PBC28411.1 QOIP01000009 RLU18831.1 KQ434893 KZC10688.1 KK107119 EZA58747.1 GEZM01001643 JAV97627.1 GBHO01044156 GBHO01036410 GBHO01010232 JAF99447.1 JAG07194.1 JAG33372.1 GBRD01018111 GDHC01020404 JAG47716.1 JAP98224.1 KB632006 ERL87962.1 APGK01034123 KB740904 ENN78450.1 KQ971342 EFA04653.1 LN849082 CRI73784.1 JH431585 GFTR01006734 JAW09692.1 KK113932 KFM61394.1 GANP01012075 JAB72393.1 ABJB010293041 DS703155 EEC05379.1 GACK01006785 JAA58249.1 PZQS01000001 PVD39003.1 GEDV01004944 JAP83613.1 KC690271 AHB53231.1 GL732530 EFX86266.1 JT416322 AHH42008.1 AJ301617 CAC37335.1 BC056280 AAH56280.2 CR407547 BC068322 AAH68322.2 KB200430 ESP01680.1 AFYH01133944 MG711509 AUT31435.1 AF336123 AAK49948.1 AERX01013317 RJVU01059775 ROJ66271.1 JARO02005224 KPP67133.1 BX927225 AF437316 AF336124 AF510101 AAL87635.1 AAM00192.1 AAN03844.1 GG666505 EEN61413.1 AF159106 BC042228 AAH42228.1 RJVU01051519 ROL41742.1 HADW01019494 SBP20894.1 AHAT01034647 CP026243 AWO97156.1 AFYH01065303 AHAT01004650 CM004482 OCT63845.1 AF039215 AJ243323 AF179213 AF114151 BC049397 AAH49397.1 CM001264 EHH21587.1 AQIA01015569 AC149036 CR759995 BC090579 AAH90579.1 AAZ06130.1 CAJ82817.1 RJVU01051648 ROL41476.1 AERX01048603 AAMC01057529 AEMK02000100 DQIR01145990 HDB01467.1 AANG04002752 AEYP01060373 CM004483 OCT61339.1 GFFV01001978 JAV37967.1 AAQR03032755
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000027135
UP000078541
+ More
UP000002358 UP000008237 UP000000311 UP000242457 UP000005203 UP000279307 UP000076502 UP000053097 UP000030742 UP000019118 UP000007266 UP000054359 UP000001555 UP000192223 UP000245119 UP000000305 UP000221080 UP000000437 UP000030746 UP000265140 UP000008672 UP000005207 UP000034805 UP000261440 UP000085678 UP000001554 UP000018468 UP000246464 UP000186698 UP000192224 UP000265160 UP000261460 UP000018467 UP000265100 UP000261560 UP000233120 UP000233100 UP000008143 UP000189706 UP000233080 UP000008227 UP000265300 UP000011712 UP000000715 UP000087266 UP000252040 UP000005225 UP000248483 UP000261400 UP000001038
UP000002358 UP000008237 UP000000311 UP000242457 UP000005203 UP000279307 UP000076502 UP000053097 UP000030742 UP000019118 UP000007266 UP000054359 UP000001555 UP000192223 UP000245119 UP000000305 UP000221080 UP000000437 UP000030746 UP000265140 UP000008672 UP000005207 UP000034805 UP000261440 UP000085678 UP000001554 UP000018468 UP000246464 UP000186698 UP000192224 UP000265160 UP000261460 UP000018467 UP000265100 UP000261560 UP000233120 UP000233100 UP000008143 UP000189706 UP000233080 UP000008227 UP000265300 UP000011712 UP000000715 UP000087266 UP000252040 UP000005225 UP000248483 UP000261400 UP000001038
PRIDE
Interpro
SUPFAM
SSF63501
SSF63501
Gene 3D
ProteinModelPortal
H9J8U3
A0A2W1BZZ7
A0A2A4K442
A0A2H1VHU6
A0A1E1WUB8
A0A194RGX7
+ More
A0A0L7LLG7 A0A1B6FSU1 A0A067QHH7 A0A1B6DUK5 A0A195FKC0 K7IXM9 E2B9L1 E2AHT4 A0A023F262 A0A2A3EBH9 A0A088A6V2 A0A3L8DES9 A0A154PH96 A0A026WRU5 A0A1Y1NNJ7 A0A0A9VZ38 A0A0K8S350 U4U4I1 N6U9T1 D6WJY3 A0A143VP60 T1JLV3 A0A224XM69 A0A087T8F5 V5HFW8 B7PFK7 L7M581 A0A1W4XSY0 A0A2T7Q003 A0A131YWN6 V5SQJ2 E9G2J9 W5UR01 A0A2D0S5B5 Q98SI2 Q7SZR7 Q6NV44 V4CHR5 A0A3P8YED9 H3AZ03 A0A2K9RCG5 Q90ZT3 I3KZU0 A0A3N0XUZ5 A0A0P7TZS4 A0A3B4CXR7 Q8QFM3 A0A1S3JF10 C3YEA5 Q8AVJ9 A0A3P8ZXJ8 A0A3N0Y6C7 A0A1A7XST0 W5NMT4 A0A2U9AZV8 H3B255 W5MX40 A0A1L8EX17 A0A1W4Y9D2 A0A3P9C180 A0A3P9D6V3 Q9PUK8 A0A3B4FUC1 A0A3B4GD67 Q7ZU08 W5LU60 A0A3P8NK42 G7N8N7 A0A1W5B4Z9 A0A3B3CTI6 A0A2K6B3T9 A0A2K5URA6 Q5BL72 A0A3N0Y5I0 A0A1W4ZY33 A0A3P8QM51 A0A1U7QEK4 A0A2K5JJR2 I3JSE2 G1K3F4 F1SI18 A0A340Y6D5 A0A2I2U7V2 M3Z5C2 A0A1L8EPZ4 A0A1S3MVZ0 A0A250Y2V1 A0A341CGM7 H0XWH9 A0A2Y9N1H2 A0A3B4ZM92 H2MKU6 A0A1S3MQT5
A0A0L7LLG7 A0A1B6FSU1 A0A067QHH7 A0A1B6DUK5 A0A195FKC0 K7IXM9 E2B9L1 E2AHT4 A0A023F262 A0A2A3EBH9 A0A088A6V2 A0A3L8DES9 A0A154PH96 A0A026WRU5 A0A1Y1NNJ7 A0A0A9VZ38 A0A0K8S350 U4U4I1 N6U9T1 D6WJY3 A0A143VP60 T1JLV3 A0A224XM69 A0A087T8F5 V5HFW8 B7PFK7 L7M581 A0A1W4XSY0 A0A2T7Q003 A0A131YWN6 V5SQJ2 E9G2J9 W5UR01 A0A2D0S5B5 Q98SI2 Q7SZR7 Q6NV44 V4CHR5 A0A3P8YED9 H3AZ03 A0A2K9RCG5 Q90ZT3 I3KZU0 A0A3N0XUZ5 A0A0P7TZS4 A0A3B4CXR7 Q8QFM3 A0A1S3JF10 C3YEA5 Q8AVJ9 A0A3P8ZXJ8 A0A3N0Y6C7 A0A1A7XST0 W5NMT4 A0A2U9AZV8 H3B255 W5MX40 A0A1L8EX17 A0A1W4Y9D2 A0A3P9C180 A0A3P9D6V3 Q9PUK8 A0A3B4FUC1 A0A3B4GD67 Q7ZU08 W5LU60 A0A3P8NK42 G7N8N7 A0A1W5B4Z9 A0A3B3CTI6 A0A2K6B3T9 A0A2K5URA6 Q5BL72 A0A3N0Y5I0 A0A1W4ZY33 A0A3P8QM51 A0A1U7QEK4 A0A2K5JJR2 I3JSE2 G1K3F4 F1SI18 A0A340Y6D5 A0A2I2U7V2 M3Z5C2 A0A1L8EPZ4 A0A1S3MVZ0 A0A250Y2V1 A0A341CGM7 H0XWH9 A0A2Y9N1H2 A0A3B4ZM92 H2MKU6 A0A1S3MQT5
PDB
6BD4
E-value=3.52885e-86,
Score=813
Ontologies
PATHWAY
GO
GO:0007275
GO:0016021
GO:0042813
GO:0016055
GO:0017147
GO:0005886
GO:0035567
GO:0060070
GO:0090504
GO:0042074
GO:0060027
GO:0035988
GO:0004930
GO:0008104
GO:0001707
GO:0045545
GO:0009950
GO:0048729
GO:0010008
GO:0045944
GO:0031018
GO:0016477
GO:0061341
GO:0060271
GO:0001947
GO:0003146
GO:0007223
GO:0070654
GO:0060231
GO:0045893
GO:0055038
GO:0014834
GO:0005546
GO:0042327
GO:2000726
GO:0030165
GO:0048103
GO:0042666
GO:0033077
GO:0038031
GO:0019827
GO:0060828
GO:0010812
GO:0005109
GO:0034446
GO:0060054
GO:0005515
GO:0004888
GO:0007166
GO:0016020
GO:0003676
GO:0043401
GO:0003677
GO:0003707
GO:0004651
GO:0005634
Topology
Subcellular location
Cell membrane
Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2). Localized in recycling endosomes in other conditions. With evidence from 2 publications.
Endosome membrane Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2). Localized in recycling endosomes in other conditions. With evidence from 2 publications.
Endosome membrane Associated to the plasma membrane in the presence of FZD7 and phosphatidylinositol 4,5-bisphosphate (PIP2). Localized in recycling endosomes in other conditions. With evidence from 2 publications.
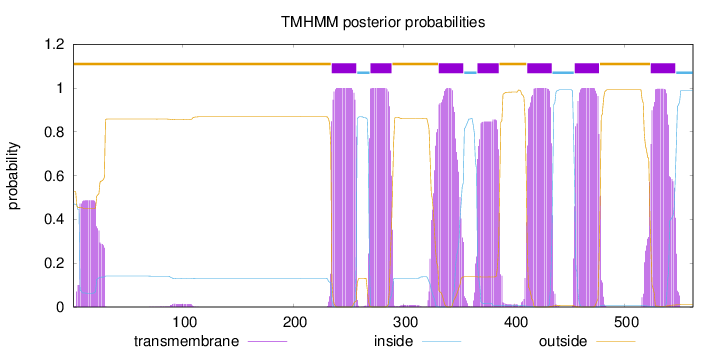
SignalP
Position: 1 - 23,
Likelihood: 0.989725

Length:
562
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
163.3181
Exp number, first 60 AAs:
10.03284
Total prob of N-in:
0.47247
POSSIBLE N-term signal
sequence
outside
1 - 234
TMhelix
235 - 257
inside
258 - 269
TMhelix
270 - 289
outside
290 - 331
TMhelix
332 - 354
inside
355 - 366
TMhelix
367 - 386
outside
387 - 411
TMhelix
412 - 434
inside
435 - 454
TMhelix
455 - 477
outside
478 - 523
TMhelix
524 - 546
inside
547 - 562
Population Genetic Test Statistics
Pi
140.565563
Theta
178.964311
Tajima's D
-0.874882
CLR
1184.20175
CSRT
0.161941902904855
Interpretation
Uncertain
