Gene
KWMTBOMO01918 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006209
Annotation
transport_protein_Sec61_beta_subunit_[Bombyx_mori]
Full name
Protein transport protein Sec61 subunit beta
+ More
DNA ligase
DNA ligase
Location in the cell
Nuclear Reliability : 3.505
Sequence
CDS
ATGCCTCAATCTCCGAGTGCTACCTCTGTAGGATCTGGGAGTAGATCACCCACAAAAGCTTCTGCTGGCCCACGAACAGCCAGTGGTAGTACAGTCCGTCAACGAAAATCAACTACTACTACAACTGCAGCCAGAAACCGAAGCACTGGTGCTGGTTCTGGAGGAATGTGGAGGTTCTATACCGATGATTCTCCTGGAGTCAAAGTGGGGCCAGTTCCTGTTTTAGTGATGTCTCTCCTGTTTATTGCATCGGTATTCATGTTACACATTTGGGGAAAATATACTAGAGCATAA
Protein
MPQSPSATSVGSGSRSPTKASAGPRTASGSTVRQRKSTTTTTAARNRSTGAGSGGMWRFYTDDSPGVKVGPVPVLVMSLLFIASVFMLHIWGKYTRA
Summary
Description
Necessary for protein translocation in the endoplasmic reticulum.
Catalytic Activity
ATP + (deoxyribonucleotide)(n)-3'-hydroxyl + 5'-phospho-(deoxyribonucleotide)(m) = (deoxyribonucleotide)(n+m) + AMP + diphosphate.
Similarity
Belongs to the SEC61-beta family.
Belongs to the ATP-dependent DNA ligase family.
Belongs to the ubiquitin-activating E1 family.
Belongs to the ATP-dependent DNA ligase family.
Belongs to the ubiquitin-activating E1 family.
Uniprot
Q2F5L5
A0A3G1T1M4
A0A0L7LLW4
I4DM80
I4DIW6
A0A2W1BUV0
+ More
A0A2J7Q7L7 S4PSG1 A0A2A4K2S8 A0A1B0GP49 A0A2H1VHW2 A0A212ETV8 A0A194RC58 A0A1B0CN43 R4V3X5 A0A1L8DTQ0 A0A067RC56 A0A224Y3A0 G3CJT4 A6YPG7 R4G497 A0A182P3S4 A0A1Y9HDT6 A0A182MWE5 A0A2M4A4Y3 R4WCV8 A0A2M3Z603 A0A2M4A504 A0A2M4C4Y1 W5JB51 A0A182FU09 A0A1I8JVE6 A0A182XC58 Q7PN12 T1E707 A0A1Q3G3H2 A0A0N7Z990 A0A182WF99 Q1HR43 Q5MIQ6 E0VEK0 A0A154PPG8 T1E3D0 A0A182YJY9 A0A2R7WL65 A0A084VSB2 A0A182MY31 T1IE84 E2B1V5 A0A182UJ46 B0WPQ9 A0A026WJZ4 A0A182G843 L7M5V1 A0A131XEU4 K7JBG6 A0A1W4X3J9 A0A195F9S6 A0A0K8TPU9 A0A0P0CW26 A0A195BYH6 A0A0B8RQ84 Q8UW24 V8NY33 A0A1W7RDK6 T1E4Y2 J3S9I4 F4WFG2 A0A158NCW1 G3MPP0 A0A224Z8H4 A0A131YEU4 A0A091G9I6 A0A1E1XM89 A0A195CR29 A0A151X624 U3FB92 A0A091I3F3 R7VQG6 A0A087QWZ6 E9IMA7 A0A093GTB3 H0ZEX2 A0A2I0MVU6 A0A2I0US50 A0A3L8T107 A0A218VB69 A0A1V4J895 A0A0Q3TND0 B5FZ77 B4LNA4 B4KMZ6 B4J786 A0A0A0AIR8 D6WLZ4 G1N714 G1KIE1 A0A091VN26 A0A091QX39 F1NWX7
A0A2J7Q7L7 S4PSG1 A0A2A4K2S8 A0A1B0GP49 A0A2H1VHW2 A0A212ETV8 A0A194RC58 A0A1B0CN43 R4V3X5 A0A1L8DTQ0 A0A067RC56 A0A224Y3A0 G3CJT4 A6YPG7 R4G497 A0A182P3S4 A0A1Y9HDT6 A0A182MWE5 A0A2M4A4Y3 R4WCV8 A0A2M3Z603 A0A2M4A504 A0A2M4C4Y1 W5JB51 A0A182FU09 A0A1I8JVE6 A0A182XC58 Q7PN12 T1E707 A0A1Q3G3H2 A0A0N7Z990 A0A182WF99 Q1HR43 Q5MIQ6 E0VEK0 A0A154PPG8 T1E3D0 A0A182YJY9 A0A2R7WL65 A0A084VSB2 A0A182MY31 T1IE84 E2B1V5 A0A182UJ46 B0WPQ9 A0A026WJZ4 A0A182G843 L7M5V1 A0A131XEU4 K7JBG6 A0A1W4X3J9 A0A195F9S6 A0A0K8TPU9 A0A0P0CW26 A0A195BYH6 A0A0B8RQ84 Q8UW24 V8NY33 A0A1W7RDK6 T1E4Y2 J3S9I4 F4WFG2 A0A158NCW1 G3MPP0 A0A224Z8H4 A0A131YEU4 A0A091G9I6 A0A1E1XM89 A0A195CR29 A0A151X624 U3FB92 A0A091I3F3 R7VQG6 A0A087QWZ6 E9IMA7 A0A093GTB3 H0ZEX2 A0A2I0MVU6 A0A2I0US50 A0A3L8T107 A0A218VB69 A0A1V4J895 A0A0Q3TND0 B5FZ77 B4LNA4 B4KMZ6 B4J786 A0A0A0AIR8 D6WLZ4 G1N714 G1KIE1 A0A091VN26 A0A091QX39 F1NWX7
EC Number
6.5.1.1
Pubmed
19121390
26227816
22651552
28756777
23622113
22118469
+ More
26354079 24845553 21058630 18207082 23691247 20920257 23761445 12364791 14747013 17210077 27129103 17204158 17510324 17244540 24945155 20566863 24330624 25244985 24438588 20798317 24508170 26483478 25576852 28049606 20075255 26369729 26445494 25476704 24297900 26358130 23758969 23025625 21719571 21347285 22216098 28797301 26830274 29209593 23915248 23371554 21282665 20360741 30282656 17018643 17994087 18362917 19820115 20838655 15592404
26354079 24845553 21058630 18207082 23691247 20920257 23761445 12364791 14747013 17210077 27129103 17204158 17510324 17244540 24945155 20566863 24330624 25244985 24438588 20798317 24508170 26483478 25576852 28049606 20075255 26369729 26445494 25476704 24297900 26358130 23758969 23025625 21719571 21347285 22216098 28797301 26830274 29209593 23915248 23371554 21282665 20360741 30282656 17018643 17994087 18362917 19820115 20838655 15592404
EMBL
BABH01005049
DQ311408
DQ631893
ABD36352.1
ABF85698.1
MG992459
+ More
AXY94897.1 JTDY01000698 KOB76176.1 AK402398 BAM19020.1 AK401234 BAM17856.1 KZ149895 PZC78828.1 NEVH01017441 PNF24578.1 GAIX01013848 JAA78712.1 NWSH01000193 PCG78561.1 AJVK01014962 ODYU01002642 SOQ40413.1 AGBW02012530 OWR44926.1 KQ460398 KPJ15187.1 AJWK01019780 KC740694 AGM32518.1 GFDF01004242 JAV09842.1 KK852561 KDR21342.1 GFTR01001445 JAW14981.1 HP639851 AEM97988.1 EF638978 GEMB01003747 ABR27863.1 JAR99473.1 ACPB03007951 GAHY01000951 JAA76559.1 AXCM01000326 GGFK01002490 MBW35811.1 AK417208 BAN20423.1 GGFM01003198 MBW23949.1 GGFK01002481 MBW35802.1 GGFJ01011060 MBW60201.1 ADMH02001745 ETN61226.1 AAAB01008966 EAA12929.5 GAMD01003442 JAA98148.1 GFDL01000693 JAV34352.1 GDKW01001211 JAI55384.1 DQ440251 CH478160 ABF18284.1 EAT33737.1 AY826154 GAPW01006468 AAV90726.1 JAC07130.1 DS235093 EEB11806.1 KQ434998 KZC13334.1 GALA01000470 JAA94382.1 KK854844 PTY19175.1 ATLV01015907 KE525041 KFB40856.1 ACPB03029634 GL444981 EFN60311.1 DS232028 EDS32468.1 KK107167 EZA56372.1 JXUM01151434 KQ571216 KXJ68221.1 GACK01006555 JAA58479.1 GEFH01004435 JAP64146.1 AAZX01002870 KQ981727 KYN36967.1 GDAI01001452 JAI16151.1 KP777785 ALJ10927.1 KQ976396 KYM92991.1 GBSH01002451 JAG66575.1 AF159543 AAL54898.1 AZIM01001552 ETE66472.1 GDAY02002208 JAV49224.1 GAAZ01002246 JAA95697.1 JU175844 GBEX01003062 AFJ51368.1 JAI11498.1 GL888115 EGI67213.1 ADTU01011954 JO843841 AEO35458.1 GFPF01011606 MAA22752.1 GEDV01010664 JAP77893.1 KL447915 KFO78623.1 GFAA01003066 JAU00369.1 KQ977408 KYN02947.1 KQ982490 KYQ55792.1 GAEP01001457 GBEW01000926 JAB53364.1 JAI09439.1 KL218152 KFP02747.1 KB375692 EMC82578.1 KL225961 KFM05750.1 GL764129 EFZ18374.1 KL217005 KFV72616.1 ABQF01031308 ABQF01031309 ABQF01031310 AKCR02000001 PKK33803.1 KZ505645 PKU48869.1 QUSF01000001 RLW13175.1 MUZQ01000018 OWK62971.1 LSYS01008581 OPJ68451.1 LMAW01002080 KQK81998.1 DQ214346 DQ214347 DQ214348 ACH44338.1 CH940648 EDW61056.1 CH933808 EDW09918.2 CH916367 EDW02104.1 KL871944 KGL94031.1 KQ971343 EFA03379.1 KL411190 KFR04070.1 KK805031 KFQ31489.1 AADN05000026
AXY94897.1 JTDY01000698 KOB76176.1 AK402398 BAM19020.1 AK401234 BAM17856.1 KZ149895 PZC78828.1 NEVH01017441 PNF24578.1 GAIX01013848 JAA78712.1 NWSH01000193 PCG78561.1 AJVK01014962 ODYU01002642 SOQ40413.1 AGBW02012530 OWR44926.1 KQ460398 KPJ15187.1 AJWK01019780 KC740694 AGM32518.1 GFDF01004242 JAV09842.1 KK852561 KDR21342.1 GFTR01001445 JAW14981.1 HP639851 AEM97988.1 EF638978 GEMB01003747 ABR27863.1 JAR99473.1 ACPB03007951 GAHY01000951 JAA76559.1 AXCM01000326 GGFK01002490 MBW35811.1 AK417208 BAN20423.1 GGFM01003198 MBW23949.1 GGFK01002481 MBW35802.1 GGFJ01011060 MBW60201.1 ADMH02001745 ETN61226.1 AAAB01008966 EAA12929.5 GAMD01003442 JAA98148.1 GFDL01000693 JAV34352.1 GDKW01001211 JAI55384.1 DQ440251 CH478160 ABF18284.1 EAT33737.1 AY826154 GAPW01006468 AAV90726.1 JAC07130.1 DS235093 EEB11806.1 KQ434998 KZC13334.1 GALA01000470 JAA94382.1 KK854844 PTY19175.1 ATLV01015907 KE525041 KFB40856.1 ACPB03029634 GL444981 EFN60311.1 DS232028 EDS32468.1 KK107167 EZA56372.1 JXUM01151434 KQ571216 KXJ68221.1 GACK01006555 JAA58479.1 GEFH01004435 JAP64146.1 AAZX01002870 KQ981727 KYN36967.1 GDAI01001452 JAI16151.1 KP777785 ALJ10927.1 KQ976396 KYM92991.1 GBSH01002451 JAG66575.1 AF159543 AAL54898.1 AZIM01001552 ETE66472.1 GDAY02002208 JAV49224.1 GAAZ01002246 JAA95697.1 JU175844 GBEX01003062 AFJ51368.1 JAI11498.1 GL888115 EGI67213.1 ADTU01011954 JO843841 AEO35458.1 GFPF01011606 MAA22752.1 GEDV01010664 JAP77893.1 KL447915 KFO78623.1 GFAA01003066 JAU00369.1 KQ977408 KYN02947.1 KQ982490 KYQ55792.1 GAEP01001457 GBEW01000926 JAB53364.1 JAI09439.1 KL218152 KFP02747.1 KB375692 EMC82578.1 KL225961 KFM05750.1 GL764129 EFZ18374.1 KL217005 KFV72616.1 ABQF01031308 ABQF01031309 ABQF01031310 AKCR02000001 PKK33803.1 KZ505645 PKU48869.1 QUSF01000001 RLW13175.1 MUZQ01000018 OWK62971.1 LSYS01008581 OPJ68451.1 LMAW01002080 KQK81998.1 DQ214346 DQ214347 DQ214348 ACH44338.1 CH940648 EDW61056.1 CH933808 EDW09918.2 CH916367 EDW02104.1 KL871944 KGL94031.1 KQ971343 EFA03379.1 KL411190 KFR04070.1 KK805031 KFQ31489.1 AADN05000026
Proteomes
UP000005204
UP000037510
UP000235965
UP000218220
UP000092462
UP000007151
+ More
UP000053240 UP000092461 UP000027135 UP000015103 UP000075885 UP000075900 UP000075883 UP000000673 UP000069272 UP000075903 UP000076407 UP000007062 UP000075920 UP000008820 UP000009046 UP000076502 UP000076408 UP000030765 UP000075884 UP000000311 UP000075902 UP000002320 UP000053097 UP000069940 UP000249989 UP000002358 UP000192223 UP000078541 UP000078540 UP000007755 UP000005205 UP000053760 UP000078542 UP000075809 UP000054308 UP000053286 UP000053875 UP000007754 UP000053872 UP000276834 UP000197619 UP000190648 UP000051836 UP000008792 UP000009192 UP000001070 UP000053858 UP000007266 UP000001645 UP000001646 UP000053283 UP000000539
UP000053240 UP000092461 UP000027135 UP000015103 UP000075885 UP000075900 UP000075883 UP000000673 UP000069272 UP000075903 UP000076407 UP000007062 UP000075920 UP000008820 UP000009046 UP000076502 UP000076408 UP000030765 UP000075884 UP000000311 UP000075902 UP000002320 UP000053097 UP000069940 UP000249989 UP000002358 UP000192223 UP000078541 UP000078540 UP000007755 UP000005205 UP000053760 UP000078542 UP000075809 UP000054308 UP000053286 UP000053875 UP000007754 UP000053872 UP000276834 UP000197619 UP000190648 UP000051836 UP000008792 UP000009192 UP000001070 UP000053858 UP000007266 UP000001645 UP000001646 UP000053283 UP000000539
PRIDE
Pfam
Interpro
IPR030671
Sec61-beta/Sbh
+ More
IPR016482 SecG/Sec61-beta/Sbh
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR042302 E1_FCCH_sf
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR032418 E1_FCCH
IPR042063 Ubi_acti_E1_SCCH
IPR018074 UBQ-activ_enz_E1_CS
IPR016059 DNA_ligase_ATP-dep_CS
IPR012340 NA-bd_OB-fold
IPR032420 E1_4HB
IPR012310 DNA_ligase_ATP-dep_cent
IPR035985 Ubiquitin-activating_enz
IPR018075 UBQ-activ_enz_E1
IPR000594 ThiF_NAD_FAD-bd
IPR038252 UBA_E1_C_sf
IPR000977 DNA_ligase_ATP-dep
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR018965 Ub-activating_enz_E1_C
IPR019572 UBA_E1_Cys
IPR012308 DNA_ligase_ATP-dep_N
IPR036599 DNA_ligase_N_sf
IPR012309 DNA_ligase_ATP-dep_C
IPR016482 SecG/Sec61-beta/Sbh
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR042302 E1_FCCH_sf
IPR033127 UBQ-activ_enz_E1_Cys_AS
IPR032418 E1_FCCH
IPR042063 Ubi_acti_E1_SCCH
IPR018074 UBQ-activ_enz_E1_CS
IPR016059 DNA_ligase_ATP-dep_CS
IPR012340 NA-bd_OB-fold
IPR032420 E1_4HB
IPR012310 DNA_ligase_ATP-dep_cent
IPR035985 Ubiquitin-activating_enz
IPR018075 UBQ-activ_enz_E1
IPR000594 ThiF_NAD_FAD-bd
IPR038252 UBA_E1_C_sf
IPR000977 DNA_ligase_ATP-dep
IPR000011 UBQ/SUMO-activ_enz_E1-like
IPR018965 Ub-activating_enz_E1_C
IPR019572 UBA_E1_Cys
IPR012308 DNA_ligase_ATP-dep_N
IPR036599 DNA_ligase_N_sf
IPR012309 DNA_ligase_ATP-dep_C
ProteinModelPortal
Q2F5L5
A0A3G1T1M4
A0A0L7LLW4
I4DM80
I4DIW6
A0A2W1BUV0
+ More
A0A2J7Q7L7 S4PSG1 A0A2A4K2S8 A0A1B0GP49 A0A2H1VHW2 A0A212ETV8 A0A194RC58 A0A1B0CN43 R4V3X5 A0A1L8DTQ0 A0A067RC56 A0A224Y3A0 G3CJT4 A6YPG7 R4G497 A0A182P3S4 A0A1Y9HDT6 A0A182MWE5 A0A2M4A4Y3 R4WCV8 A0A2M3Z603 A0A2M4A504 A0A2M4C4Y1 W5JB51 A0A182FU09 A0A1I8JVE6 A0A182XC58 Q7PN12 T1E707 A0A1Q3G3H2 A0A0N7Z990 A0A182WF99 Q1HR43 Q5MIQ6 E0VEK0 A0A154PPG8 T1E3D0 A0A182YJY9 A0A2R7WL65 A0A084VSB2 A0A182MY31 T1IE84 E2B1V5 A0A182UJ46 B0WPQ9 A0A026WJZ4 A0A182G843 L7M5V1 A0A131XEU4 K7JBG6 A0A1W4X3J9 A0A195F9S6 A0A0K8TPU9 A0A0P0CW26 A0A195BYH6 A0A0B8RQ84 Q8UW24 V8NY33 A0A1W7RDK6 T1E4Y2 J3S9I4 F4WFG2 A0A158NCW1 G3MPP0 A0A224Z8H4 A0A131YEU4 A0A091G9I6 A0A1E1XM89 A0A195CR29 A0A151X624 U3FB92 A0A091I3F3 R7VQG6 A0A087QWZ6 E9IMA7 A0A093GTB3 H0ZEX2 A0A2I0MVU6 A0A2I0US50 A0A3L8T107 A0A218VB69 A0A1V4J895 A0A0Q3TND0 B5FZ77 B4LNA4 B4KMZ6 B4J786 A0A0A0AIR8 D6WLZ4 G1N714 G1KIE1 A0A091VN26 A0A091QX39 F1NWX7
A0A2J7Q7L7 S4PSG1 A0A2A4K2S8 A0A1B0GP49 A0A2H1VHW2 A0A212ETV8 A0A194RC58 A0A1B0CN43 R4V3X5 A0A1L8DTQ0 A0A067RC56 A0A224Y3A0 G3CJT4 A6YPG7 R4G497 A0A182P3S4 A0A1Y9HDT6 A0A182MWE5 A0A2M4A4Y3 R4WCV8 A0A2M3Z603 A0A2M4A504 A0A2M4C4Y1 W5JB51 A0A182FU09 A0A1I8JVE6 A0A182XC58 Q7PN12 T1E707 A0A1Q3G3H2 A0A0N7Z990 A0A182WF99 Q1HR43 Q5MIQ6 E0VEK0 A0A154PPG8 T1E3D0 A0A182YJY9 A0A2R7WL65 A0A084VSB2 A0A182MY31 T1IE84 E2B1V5 A0A182UJ46 B0WPQ9 A0A026WJZ4 A0A182G843 L7M5V1 A0A131XEU4 K7JBG6 A0A1W4X3J9 A0A195F9S6 A0A0K8TPU9 A0A0P0CW26 A0A195BYH6 A0A0B8RQ84 Q8UW24 V8NY33 A0A1W7RDK6 T1E4Y2 J3S9I4 F4WFG2 A0A158NCW1 G3MPP0 A0A224Z8H4 A0A131YEU4 A0A091G9I6 A0A1E1XM89 A0A195CR29 A0A151X624 U3FB92 A0A091I3F3 R7VQG6 A0A087QWZ6 E9IMA7 A0A093GTB3 H0ZEX2 A0A2I0MVU6 A0A2I0US50 A0A3L8T107 A0A218VB69 A0A1V4J895 A0A0Q3TND0 B5FZ77 B4LNA4 B4KMZ6 B4J786 A0A0A0AIR8 D6WLZ4 G1N714 G1KIE1 A0A091VN26 A0A091QX39 F1NWX7
PDB
4CG6
E-value=4.7298e-11,
Score=156
Ontologies
PATHWAY
GO
PANTHER
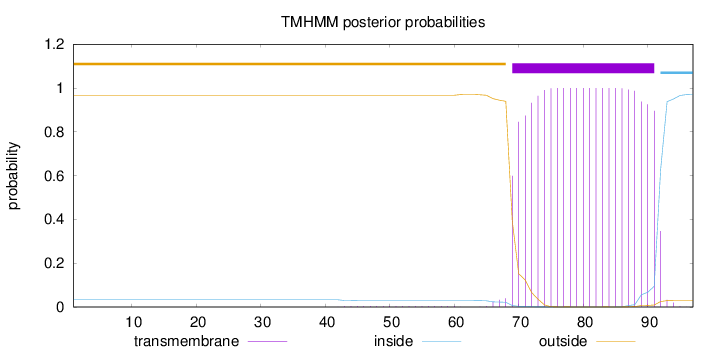
Topology
Length:
97
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.51626
Exp number, first 60 AAs:
0.06918
Total prob of N-in:
0.03316
outside
1 - 68
TMhelix
69 - 91
inside
92 - 97
Population Genetic Test Statistics
Pi
43.12894
Theta
127.960077
Tajima's D
-2.106814
CLR
800.424303
CSRT
0.00909954502274886
Interpretation
Uncertain
