Gene
KWMTBOMO01917
Pre Gene Modal
BGIBMGA005936
Annotation
PREDICTED:_2'?5'-phosphodiesterase_12_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 1.22
Sequence
CDS
ATGAGTGATGAAAATATACAGTGGGAGTTTATATGTGAAAACTTCTCCTATATTCCTACATTGAATGACATTGGAATGAAGCTTAAAGTGCAATGCATTCCAGGAAATGAAGTTACTTCAGGCCCTGCTGTTGATGCAATATCCAAAGGTGTTGTGGAAGCAGGTCCCGGCAAATGTCCTTTTGAAACAAGGCACATGTTTACAACCTCTGTTCTCAAAGGCAAAAGTTTTCGCTGTGTTTCTTATAATATTTTAGCTGATTTATATTGCGATTCTGAATTCACAAGGACTGTGCTTCATCCTTATTGCCCGCCATATGCTCTTAATATAGATTATAGAAAACAATTATTAATGAAAGAATTACTAGGATACAATGCTGATATAATATGTCTTCAAGAAGTAGATAGAAAAATATTTCAACATTGGTTGTCTCCCTTTCTTGACAATGCTGGATTTAAGGGACAGTTTTACAGAAAAGGAAAAGAGGTTGCAGAAGGACTTGCTTGTTTTTATAGATCATTAAGGTTTAGGTTCTTAGGGGAAGAACAAATAATCTTATCAGAAGCAATTAAAACTTATCCATGCTTACAGAATATATGGAATATCATAAAGACCAATGGTCCACTTGTAGAACGACTTTTAGACAGATCTACAGTAGCTAGTGTTACATTATTGGAGTCTGTAGAAAATTCGGACGAACTGCTACTTGTTGGAAATACACATTTGTATTTCCACCCCGATGCTGATCACATAAGACTTATTCAAGGTGGCATTTTTATTTATTGGATAAACGATGTTAGGAAAAAAATTATGGAAAAGTTTCCCGATAAAAAAATTAGCTTGATTTTGTGCGGCGATTATAACAGTGTACCAACTTGCGGAATATACCAATTGTATACAACAGGAATAGCACCAAGTACATTGCCAGATTGGAAATCCAATGAGACCGAAAGTGTACAAGGTTTGAATCTGGAGCAGGATATCTTACTGGCCAGTGCATGTGGTACACCAACATACACAAATTTTACTGCTGAATTTGCTGATTGTTTGGATTATATATTTTACGACACATCCAGTTTAGAGGTTGAACAGGTAATTCCGTTCCCGAGTATAGATGAGTTAAAAGCCAACGTTGCCTTACCAAGTGTTGTTTTTCCATCGGACCACATAGCTTTAATATCAGATTTACGATTCAAATGA
Protein
MSDENIQWEFICENFSYIPTLNDIGMKLKVQCIPGNEVTSGPAVDAISKGVVEAGPGKCPFETRHMFTTSVLKGKSFRCVSYNILADLYCDSEFTRTVLHPYCPPYALNIDYRKQLLMKELLGYNADIICLQEVDRKIFQHWLSPFLDNAGFKGQFYRKGKEVAEGLACFYRSLRFRFLGEEQIILSEAIKTYPCLQNIWNIIKTNGPLVERLLDRSTVASVTLLESVENSDELLLVGNTHLYFHPDADHIRLIQGGIFIYWINDVRKKIMEKFPDKKISLILCGDYNSVPTCGIYQLYTTGIAPSTLPDWKSNETESVQGLNLEQDILLASACGTPTYTNFTAEFADCLDYIFYDTSSLEVEQVIPFPSIDELKANVALPSVVFPSDHIALISDLRFK
Summary
Uniprot
H9J8U4
A0A2W1BV32
A0A2A4K4H9
A0A194RBI0
A0A194PRV7
A0A212ETZ2
+ More
A0A0L7LLW4 A0A2H1VJ14 N6UD15 D6WJI3 A0A067QJY3 A0A1W4WGE4 A0A1I8N4Y1 A0A158P3P0 A0A1Y1K9A4 E9IK67 A0A336LEE4 E2ACV1 A0A195BY22 F4X151 A0A195CSZ5 A0A0L0BUV2 A0A1I8PS33 A0A195F1B8 A0A1A9WA61 A0A0K8TKM9 A0A026WF76 A0A151XG65 A0A1B6D1I8 A0A0A1WP40 A0A0K8UKA8 A0A2J7QF48 A0A0K8W382 A0A1B6L0R0 T1HYV8 A0A0K8UCB7 A0A0K8W490 A0A1B0C8X0 A0A151JPP6 A0A034WC48 A0A1B6K4P8 A0A1A9XXZ6 U4U2M8 W8CD91 D3TNH5 A0A1B0G5S0 B0XJI2 E2C449 A0A1B0BX14 A0A1A9Z8J2 A0A182GSR5 E9HCR1 A0A182PDF4 A0A1Z5LH20 A0A0P6IV41 Q0IFM1 A0A182FIP8 A0A1S4F8I6 A0A0P5U0X6 A0A182IU26 A0A023FBF2 A0A0V0G8W2 A0A0P5XL80 A0A2R5LMC1 A0A084VNV8 A0A1Q3F6I5 A0A2M3ZA89 A0A182WGX1 A0A0P6FWI0 A0A1Q3F6L4 A0A2M3ZA64 A0A1Q3F6S4 A0A2M3Z9Z6 A0A0P5YDC0 Q7PW62 A0A0P5WTZ4 A0A182YFG9 A0A182KUS4 A0A182X2Y5 A0A182TYZ8 A0A069DUP6 A0A2M4AJ57 A0A1S4H0V0 A0A224XP01 A0A182V6S5 A0A2C9GRW1 A0A0P6GFQ5 A0A182K3Q0 A0A2M4CRN5 A0A2M4CRH8 A0A0N8DE82 A0A2M4BII6 A0A1B6FFQ0 A0A182R9S1 A0A1J1HMZ7 A0A182ML47 A0A293MPY6 A0A0P4Z8I7 A0A0P5TSA3 A0A0P8XFX8
A0A0L7LLW4 A0A2H1VJ14 N6UD15 D6WJI3 A0A067QJY3 A0A1W4WGE4 A0A1I8N4Y1 A0A158P3P0 A0A1Y1K9A4 E9IK67 A0A336LEE4 E2ACV1 A0A195BY22 F4X151 A0A195CSZ5 A0A0L0BUV2 A0A1I8PS33 A0A195F1B8 A0A1A9WA61 A0A0K8TKM9 A0A026WF76 A0A151XG65 A0A1B6D1I8 A0A0A1WP40 A0A0K8UKA8 A0A2J7QF48 A0A0K8W382 A0A1B6L0R0 T1HYV8 A0A0K8UCB7 A0A0K8W490 A0A1B0C8X0 A0A151JPP6 A0A034WC48 A0A1B6K4P8 A0A1A9XXZ6 U4U2M8 W8CD91 D3TNH5 A0A1B0G5S0 B0XJI2 E2C449 A0A1B0BX14 A0A1A9Z8J2 A0A182GSR5 E9HCR1 A0A182PDF4 A0A1Z5LH20 A0A0P6IV41 Q0IFM1 A0A182FIP8 A0A1S4F8I6 A0A0P5U0X6 A0A182IU26 A0A023FBF2 A0A0V0G8W2 A0A0P5XL80 A0A2R5LMC1 A0A084VNV8 A0A1Q3F6I5 A0A2M3ZA89 A0A182WGX1 A0A0P6FWI0 A0A1Q3F6L4 A0A2M3ZA64 A0A1Q3F6S4 A0A2M3Z9Z6 A0A0P5YDC0 Q7PW62 A0A0P5WTZ4 A0A182YFG9 A0A182KUS4 A0A182X2Y5 A0A182TYZ8 A0A069DUP6 A0A2M4AJ57 A0A1S4H0V0 A0A224XP01 A0A182V6S5 A0A2C9GRW1 A0A0P6GFQ5 A0A182K3Q0 A0A2M4CRN5 A0A2M4CRH8 A0A0N8DE82 A0A2M4BII6 A0A1B6FFQ0 A0A182R9S1 A0A1J1HMZ7 A0A182ML47 A0A293MPY6 A0A0P4Z8I7 A0A0P5TSA3 A0A0P8XFX8
Pubmed
19121390
28756777
26354079
22118469
26227816
23537049
+ More
18362917 19820115 24845553 25315136 21347285 28004739 21282665 20798317 21719571 26108605 26369729 24508170 30249741 25830018 25348373 24495485 20353571 26483478 21292972 28528879 26999592 17510324 25474469 24438588 12364791 25244985 20966253 26334808 17994087 18057021
18362917 19820115 24845553 25315136 21347285 28004739 21282665 20798317 21719571 26108605 26369729 24508170 30249741 25830018 25348373 24495485 20353571 26483478 21292972 28528879 26999592 17510324 25474469 24438588 12364791 25244985 20966253 26334808 17994087 18057021
EMBL
BABH01005048
BABH01005049
KZ149895
PZC78829.1
NWSH01000193
PCG78560.1
+ More
KQ460398 KPJ15188.1 KQ459601 KPI93870.1 AGBW02012530 OWR44927.1 JTDY01000698 KOB76176.1 ODYU01002642 SOQ40412.1 APGK01028352 KB740686 ENN79560.1 KQ971342 EFA03899.1 KK853378 KDR08025.1 ADTU01008307 GEZM01091241 JAV56998.1 GL763924 EFZ19036.1 UFQS01004139 UFQT01004139 SSX16206.1 SSX35532.1 GL438582 EFN68749.1 KQ976394 KYM93195.1 GL888526 EGI59818.1 KQ977304 KYN03823.1 JRES01001295 KNC23845.1 KQ981880 KYN33962.1 GDAI01002915 JAI14688.1 KK107250 QOIP01000002 EZA54336.1 RLU25525.1 KQ982174 KYQ59366.1 GEDC01017863 JAS19435.1 GBXI01014099 GBXI01005346 JAD00193.1 JAD08946.1 GDHF01025167 JAI27147.1 NEVH01015302 PNF27206.1 GDHF01006785 JAI45529.1 GEBQ01022695 JAT17282.1 ACPB03017078 GDHF01028007 JAI24307.1 GDHF01006276 JAI46038.1 AJWK01001553 KQ978781 KYN28485.1 GAKP01007579 JAC51373.1 GECU01001289 JAT06418.1 KB631930 ERL87317.1 GAMC01002356 JAC04200.1 EZ422977 ADD19253.1 CCAG010015749 CCAG010015750 DS233522 EDS30409.1 GL452396 EFN77278.1 JXJN01022027 JXUM01085309 KQ563494 KXJ73751.1 GL732621 EFX70391.1 GFJQ02000291 JAW06679.1 GDUN01000329 JAN95590.1 CH477312 EAT43882.1 GDIP01119338 JAL84376.1 GBBI01000022 JAC18690.1 GECL01001747 JAP04377.1 GDIP01070403 JAM33312.1 GGLE01006452 MBY10578.1 ATLV01014931 KE524996 KFB39652.1 GFDL01011864 JAV23181.1 GGFM01004695 MBW25446.1 GDIQ01041810 JAN52927.1 GFDL01011853 JAV23192.1 GGFM01004577 MBW25328.1 GFDL01011867 JAV23178.1 GGFM01004549 MBW25300.1 GDIP01059296 LRGB01000446 JAM44419.1 KZS19075.1 AAAB01008984 EAA15055.3 GDIP01081786 JAM21929.1 GBGD01001492 JAC87397.1 GGFK01007495 MBW40816.1 GFTR01006256 JAW10170.1 APCN01003737 GDIQ01053863 JAN40874.1 GGFL01003757 MBW67935.1 GGFL01003756 MBW67934.1 GDIP01042391 JAM61324.1 GGFJ01003630 MBW52771.1 GECZ01020752 JAS49017.1 CVRI01000004 CRK87609.1 AXCM01001875 GFWV01023000 MAA47727.1 GDIP01216274 JAJ07128.1 GDIP01127681 JAL76033.1 CH902620 KPU73813.1 KPU73814.1
KQ460398 KPJ15188.1 KQ459601 KPI93870.1 AGBW02012530 OWR44927.1 JTDY01000698 KOB76176.1 ODYU01002642 SOQ40412.1 APGK01028352 KB740686 ENN79560.1 KQ971342 EFA03899.1 KK853378 KDR08025.1 ADTU01008307 GEZM01091241 JAV56998.1 GL763924 EFZ19036.1 UFQS01004139 UFQT01004139 SSX16206.1 SSX35532.1 GL438582 EFN68749.1 KQ976394 KYM93195.1 GL888526 EGI59818.1 KQ977304 KYN03823.1 JRES01001295 KNC23845.1 KQ981880 KYN33962.1 GDAI01002915 JAI14688.1 KK107250 QOIP01000002 EZA54336.1 RLU25525.1 KQ982174 KYQ59366.1 GEDC01017863 JAS19435.1 GBXI01014099 GBXI01005346 JAD00193.1 JAD08946.1 GDHF01025167 JAI27147.1 NEVH01015302 PNF27206.1 GDHF01006785 JAI45529.1 GEBQ01022695 JAT17282.1 ACPB03017078 GDHF01028007 JAI24307.1 GDHF01006276 JAI46038.1 AJWK01001553 KQ978781 KYN28485.1 GAKP01007579 JAC51373.1 GECU01001289 JAT06418.1 KB631930 ERL87317.1 GAMC01002356 JAC04200.1 EZ422977 ADD19253.1 CCAG010015749 CCAG010015750 DS233522 EDS30409.1 GL452396 EFN77278.1 JXJN01022027 JXUM01085309 KQ563494 KXJ73751.1 GL732621 EFX70391.1 GFJQ02000291 JAW06679.1 GDUN01000329 JAN95590.1 CH477312 EAT43882.1 GDIP01119338 JAL84376.1 GBBI01000022 JAC18690.1 GECL01001747 JAP04377.1 GDIP01070403 JAM33312.1 GGLE01006452 MBY10578.1 ATLV01014931 KE524996 KFB39652.1 GFDL01011864 JAV23181.1 GGFM01004695 MBW25446.1 GDIQ01041810 JAN52927.1 GFDL01011853 JAV23192.1 GGFM01004577 MBW25328.1 GFDL01011867 JAV23178.1 GGFM01004549 MBW25300.1 GDIP01059296 LRGB01000446 JAM44419.1 KZS19075.1 AAAB01008984 EAA15055.3 GDIP01081786 JAM21929.1 GBGD01001492 JAC87397.1 GGFK01007495 MBW40816.1 GFTR01006256 JAW10170.1 APCN01003737 GDIQ01053863 JAN40874.1 GGFL01003757 MBW67935.1 GGFL01003756 MBW67934.1 GDIP01042391 JAM61324.1 GGFJ01003630 MBW52771.1 GECZ01020752 JAS49017.1 CVRI01000004 CRK87609.1 AXCM01001875 GFWV01023000 MAA47727.1 GDIP01216274 JAJ07128.1 GDIP01127681 JAL76033.1 CH902620 KPU73813.1 KPU73814.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000019118 UP000007266 UP000027135 UP000192223 UP000095301 UP000005205 UP000000311 UP000078540 UP000007755 UP000078542 UP000037069 UP000095300 UP000078541 UP000091820 UP000053097 UP000279307 UP000075809 UP000235965 UP000015103 UP000092461 UP000078492 UP000092443 UP000030742 UP000092444 UP000002320 UP000008237 UP000092460 UP000092445 UP000069940 UP000249989 UP000000305 UP000075885 UP000008820 UP000069272 UP000075880 UP000030765 UP000075920 UP000076858 UP000007062 UP000076408 UP000075882 UP000076407 UP000075902 UP000075903 UP000075840 UP000075881 UP000075900 UP000183832 UP000075883 UP000007801
UP000019118 UP000007266 UP000027135 UP000192223 UP000095301 UP000005205 UP000000311 UP000078540 UP000007755 UP000078542 UP000037069 UP000095300 UP000078541 UP000091820 UP000053097 UP000279307 UP000075809 UP000235965 UP000015103 UP000092461 UP000078492 UP000092443 UP000030742 UP000092444 UP000002320 UP000008237 UP000092460 UP000092445 UP000069940 UP000249989 UP000000305 UP000075885 UP000008820 UP000069272 UP000075880 UP000030765 UP000075920 UP000076858 UP000007062 UP000076408 UP000075882 UP000076407 UP000075902 UP000075903 UP000075840 UP000075881 UP000075900 UP000183832 UP000075883 UP000007801
PRIDE
Pfam
Interpro
IPR005135
Endo/exonuclease/phosphatase
+ More
IPR036691 Endo/exonu/phosph_ase_sf
IPR016482 SecG/Sec61-beta/Sbh
IPR000595 cNMP-bd_dom
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR035014 STKc_cGK
IPR014710 RmlC-like_jellyroll
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR018490 cNMP-bd-like
IPR017441 Protein_kinase_ATP_BS
IPR002809 EMC3/TMCO1
IPR036691 Endo/exonu/phosph_ase_sf
IPR016482 SecG/Sec61-beta/Sbh
IPR000595 cNMP-bd_dom
IPR000961 AGC-kinase_C
IPR000719 Prot_kinase_dom
IPR035014 STKc_cGK
IPR014710 RmlC-like_jellyroll
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR018490 cNMP-bd-like
IPR017441 Protein_kinase_ATP_BS
IPR002809 EMC3/TMCO1
Gene 3D
ProteinModelPortal
H9J8U4
A0A2W1BV32
A0A2A4K4H9
A0A194RBI0
A0A194PRV7
A0A212ETZ2
+ More
A0A0L7LLW4 A0A2H1VJ14 N6UD15 D6WJI3 A0A067QJY3 A0A1W4WGE4 A0A1I8N4Y1 A0A158P3P0 A0A1Y1K9A4 E9IK67 A0A336LEE4 E2ACV1 A0A195BY22 F4X151 A0A195CSZ5 A0A0L0BUV2 A0A1I8PS33 A0A195F1B8 A0A1A9WA61 A0A0K8TKM9 A0A026WF76 A0A151XG65 A0A1B6D1I8 A0A0A1WP40 A0A0K8UKA8 A0A2J7QF48 A0A0K8W382 A0A1B6L0R0 T1HYV8 A0A0K8UCB7 A0A0K8W490 A0A1B0C8X0 A0A151JPP6 A0A034WC48 A0A1B6K4P8 A0A1A9XXZ6 U4U2M8 W8CD91 D3TNH5 A0A1B0G5S0 B0XJI2 E2C449 A0A1B0BX14 A0A1A9Z8J2 A0A182GSR5 E9HCR1 A0A182PDF4 A0A1Z5LH20 A0A0P6IV41 Q0IFM1 A0A182FIP8 A0A1S4F8I6 A0A0P5U0X6 A0A182IU26 A0A023FBF2 A0A0V0G8W2 A0A0P5XL80 A0A2R5LMC1 A0A084VNV8 A0A1Q3F6I5 A0A2M3ZA89 A0A182WGX1 A0A0P6FWI0 A0A1Q3F6L4 A0A2M3ZA64 A0A1Q3F6S4 A0A2M3Z9Z6 A0A0P5YDC0 Q7PW62 A0A0P5WTZ4 A0A182YFG9 A0A182KUS4 A0A182X2Y5 A0A182TYZ8 A0A069DUP6 A0A2M4AJ57 A0A1S4H0V0 A0A224XP01 A0A182V6S5 A0A2C9GRW1 A0A0P6GFQ5 A0A182K3Q0 A0A2M4CRN5 A0A2M4CRH8 A0A0N8DE82 A0A2M4BII6 A0A1B6FFQ0 A0A182R9S1 A0A1J1HMZ7 A0A182ML47 A0A293MPY6 A0A0P4Z8I7 A0A0P5TSA3 A0A0P8XFX8
A0A0L7LLW4 A0A2H1VJ14 N6UD15 D6WJI3 A0A067QJY3 A0A1W4WGE4 A0A1I8N4Y1 A0A158P3P0 A0A1Y1K9A4 E9IK67 A0A336LEE4 E2ACV1 A0A195BY22 F4X151 A0A195CSZ5 A0A0L0BUV2 A0A1I8PS33 A0A195F1B8 A0A1A9WA61 A0A0K8TKM9 A0A026WF76 A0A151XG65 A0A1B6D1I8 A0A0A1WP40 A0A0K8UKA8 A0A2J7QF48 A0A0K8W382 A0A1B6L0R0 T1HYV8 A0A0K8UCB7 A0A0K8W490 A0A1B0C8X0 A0A151JPP6 A0A034WC48 A0A1B6K4P8 A0A1A9XXZ6 U4U2M8 W8CD91 D3TNH5 A0A1B0G5S0 B0XJI2 E2C449 A0A1B0BX14 A0A1A9Z8J2 A0A182GSR5 E9HCR1 A0A182PDF4 A0A1Z5LH20 A0A0P6IV41 Q0IFM1 A0A182FIP8 A0A1S4F8I6 A0A0P5U0X6 A0A182IU26 A0A023FBF2 A0A0V0G8W2 A0A0P5XL80 A0A2R5LMC1 A0A084VNV8 A0A1Q3F6I5 A0A2M3ZA89 A0A182WGX1 A0A0P6FWI0 A0A1Q3F6L4 A0A2M3ZA64 A0A1Q3F6S4 A0A2M3Z9Z6 A0A0P5YDC0 Q7PW62 A0A0P5WTZ4 A0A182YFG9 A0A182KUS4 A0A182X2Y5 A0A182TYZ8 A0A069DUP6 A0A2M4AJ57 A0A1S4H0V0 A0A224XP01 A0A182V6S5 A0A2C9GRW1 A0A0P6GFQ5 A0A182K3Q0 A0A2M4CRN5 A0A2M4CRH8 A0A0N8DE82 A0A2M4BII6 A0A1B6FFQ0 A0A182R9S1 A0A1J1HMZ7 A0A182ML47 A0A293MPY6 A0A0P4Z8I7 A0A0P5TSA3 A0A0P8XFX8
PDB
4Z0V
E-value=3.45672e-93,
Score=872
Ontologies
KEGG
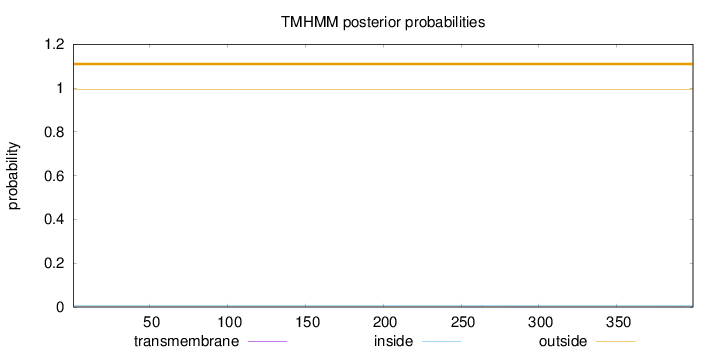
Topology
Length:
399
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01504
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00760
outside
1 - 399
Population Genetic Test Statistics
Pi
242.376585
Theta
157.084323
Tajima's D
-1.025196
CLR
0.437134
CSRT
0.130143492825359
Interpretation
Uncertain
