Gene
KWMTBOMO01916
Pre Gene Modal
BGIBMGA005937
Annotation
Uncharacterized_protein_OBRU01_01001?_partial_[Operophtera_brumata]
Location in the cell
Cytoplasmic Reliability : 1.018 Extracellular Reliability : 1.656 Nuclear Reliability : 2.765
Sequence
CDS
ATGTTCAGATTATATCTAAAGCCGAATTGCATTTCGATGGGTAAATATCAAATAGCCATTGGTAGTTCTTACATTAAACAGAATGTACACGTGTCTCCGCTATTGAAATCACAGAGTGTAGCTTTGAAATTACCCACGAATGTTGTTGATATTAACGCATCCGAAGTAGGGAGATTTTCTCCGAACAGAGAAAGAGATATTTCCGGTCCTGTTATTAATTCTCACTATAAAAAACAACATGACATGTCTAAAATTGATATAACAATACAAGTTGACCCATATGTTAACAAATATGATTGTCGAGGGGCTGTTAGCCTATCTTCGTCCATGCAGAGCAACGTTCCAAGTCCTTTCACAGGAAACCATACTCATCTCAACATGCCAGGCTCACAATGGGGACAAGGCCACAAATACTTATCAGATCATCTTCACAGTGCACATTATTTGAGTCTAAGAAATGAATACAGAGAAAAAATTATAAATAAAATTGTTGGTGTACGAAACATGAGTACGAATGTGAAGCCCCCTGTAAGTGCTAAAGAGAAACTCAAAAAGGCTGTTAAAGAGTATGGCTCCACAGTGATTGTTTTTCACGTAACAATTTCACTGATTTCATTAGGCAGTTGCTACTTGCTTGTTTCTAGTGGAGTTGATTTAGTGGCTATTCTAAAATATTTAAATATAGAAGAGGGAACTGTCTTGAAGGCTGCTGGTTCAAATGCTGGGACTTTTGTTATAGCCTATACAGTCCACAAAATTTTCGCTCCATTCCGGATAGCCATAACTTTGACTGCAACACCTTTCATCGTTCGACACTTAAGAAATATTGGAATTTTAAAACGTGGTATAAATGCTGGTGGTGGGAAATAA
Protein
MFRLYLKPNCISMGKYQIAIGSSYIKQNVHVSPLLKSQSVALKLPTNVVDINASEVGRFSPNRERDISGPVINSHYKKQHDMSKIDITIQVDPYVNKYDCRGAVSLSSSMQSNVPSPFTGNHTHLNMPGSQWGQGHKYLSDHLHSAHYLSLRNEYREKIINKIVGVRNMSTNVKPPVSAKEKLKKAVKEYGSTVIVFHVTISLISLGSCYLLVSSGVDLVAILKYLNIEEGTVLKAAGSNAGTFVIAYTVHKIFAPFRIAITLTATPFIVRHLRNIGILKRGINAGGGK
Summary
Uniprot
A0A2H1WIW1
A0A2A4K3D3
A0A2W1C137
A0A0L7LSG1
A0A194PKA6
A0A194RCJ2
+ More
A0A212ETW4 A0A1E1W6L8 A0A182PAQ5 Q7Q4K1 A0A1S4GY95 A0A182XFT5 A0A1Q3FIU6 A0A084VM95 W8AZ70 A0A034WLY2 A0A0A1WEJ1 B0XJP3 A0A0K8UVF9 A0A3B0KGT0 A0A0L0CK33 B5DJA8 A0A2P8ZE82 A0A1B0GJH4 B4GXA4 A0A1L8D9G5 A0A1L8D9T6 B4JPE6 B4MUM0 B4KI96 A0A1A9VIT8 A0A1A9Z408 A0A1A9WXH8 B4M8Z7 A0A3B0KBM8 A0A067R345 B4QCM8 A0A0M4E662 A0A2J7R0K1 B3N3K1 A0A0C9RS63 A0A1B0FEZ2 A0A336M0D1 A1Z6G4 A0A310S727 A0A1I8MQZ2 A0A195BW22 Q8SYU4 A0A158NEF7 A0A0L7QM12 A0A0N8NZY2 A0A1B0D3V6 A0A195EP58 F4WVJ6 A0A195CPV8 A0A026WSU4 A0A088A6Q4 A0A0N0U781 A0A2A3E9L5 A0A232EMG5 E2AHT3 A0A0K8WCP7 T1DGX6 B0X5Y7 E0VVE4 A0A0P5J8X8 A0A0P5FBA1 A0A0P6B3Y3 A0A0P5G5E8 A0A0P6AG28 E9FZQ8
A0A212ETW4 A0A1E1W6L8 A0A182PAQ5 Q7Q4K1 A0A1S4GY95 A0A182XFT5 A0A1Q3FIU6 A0A084VM95 W8AZ70 A0A034WLY2 A0A0A1WEJ1 B0XJP3 A0A0K8UVF9 A0A3B0KGT0 A0A0L0CK33 B5DJA8 A0A2P8ZE82 A0A1B0GJH4 B4GXA4 A0A1L8D9G5 A0A1L8D9T6 B4JPE6 B4MUM0 B4KI96 A0A1A9VIT8 A0A1A9Z408 A0A1A9WXH8 B4M8Z7 A0A3B0KBM8 A0A067R345 B4QCM8 A0A0M4E662 A0A2J7R0K1 B3N3K1 A0A0C9RS63 A0A1B0FEZ2 A0A336M0D1 A1Z6G4 A0A310S727 A0A1I8MQZ2 A0A195BW22 Q8SYU4 A0A158NEF7 A0A0L7QM12 A0A0N8NZY2 A0A1B0D3V6 A0A195EP58 F4WVJ6 A0A195CPV8 A0A026WSU4 A0A088A6Q4 A0A0N0U781 A0A2A3E9L5 A0A232EMG5 E2AHT3 A0A0K8WCP7 T1DGX6 B0X5Y7 E0VVE4 A0A0P5J8X8 A0A0P5FBA1 A0A0P6B3Y3 A0A0P5G5E8 A0A0P6AG28 E9FZQ8
Pubmed
28756777
26227816
26354079
22118469
12364791
24438588
+ More
24495485 25348373 25830018 26108605 15632085 17994087 29403074 24845553 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 21347285 21719571 24508170 28648823 20798317 24330624 20566863 21292972
24495485 25348373 25830018 26108605 15632085 17994087 29403074 24845553 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 21347285 21719571 24508170 28648823 20798317 24330624 20566863 21292972
EMBL
ODYU01008632
SOQ52384.1
NWSH01000193
PCG78559.1
KZ149895
PZC78830.1
+ More
JTDY01000181 KOB78413.1 KQ459601 KPI93871.1 KQ460398 KPJ15189.1 AGBW02012530 OWR44928.1 GDQN01008412 JAT82642.1 AAAB01008964 EAA12719.3 GFDL01007600 JAV27445.1 ATLV01014582 KE524975 KFB39089.1 GAMC01015023 JAB91532.1 GAKP01003585 JAC55367.1 GBXI01017000 JAC97291.1 DS233573 EDS30582.1 GDHF01021657 JAI30657.1 OUUW01000010 SPP85569.1 JRES01000300 KNC32610.1 CH379061 EDY70677.2 PYGN01000083 PSN54798.1 AJWK01021695 AJWK01021696 AJWK01021697 AJWK01021698 CH479195 EDW27215.1 GFDF01010958 JAV03126.1 GFDF01010959 JAV03125.1 CH916372 EDV98776.1 CH963857 EDW76215.1 CH933807 EDW12389.2 CH940654 EDW57673.2 SPP85570.1 KK853038 KDR12191.1 CM000362 CM002911 EDX05817.1 KMY91630.1 CP012523 ALC38015.1 NEVH01008240 PNF34346.1 CH954177 EDV59883.2 GBYB01011464 GBYB01011465 GBYB01011466 JAG81231.1 JAG81232.1 JAG81233.1 CCAG010000397 UFQS01000381 UFQT01000381 SSX03421.1 SSX23786.1 AE013599 BT128906 AAM68369.1 AEO72333.1 KQ776753 OAD52186.1 KQ976401 KYM92480.1 AY071311 AAL48933.1 ADTU01013285 KQ414905 KOC59604.1 CH902638 KPU75596.1 AJVK01011049 AJVK01011050 AJVK01011051 KQ978625 KYN29669.1 GL888387 EGI61817.1 KQ977444 KYN02665.1 KK107119 EZA58746.1 KQ435709 KOX80052.1 KZ288312 PBC28410.1 NNAY01003371 OXU19531.1 GL439579 EFN67012.1 GDHF01003457 JAI48857.1 GALA01001496 JAA93356.1 DS232397 EDS41118.1 DS235811 EEB17350.1 GDIQ01202975 GDIP01091067 JAK48750.1 JAM12648.1 GDIQ01256924 JAJ94800.1 GDIP01020372 JAM83343.1 GDIQ01250804 JAK00921.1 GDIP01043218 JAM60497.1 GL732528 EFX87100.1
JTDY01000181 KOB78413.1 KQ459601 KPI93871.1 KQ460398 KPJ15189.1 AGBW02012530 OWR44928.1 GDQN01008412 JAT82642.1 AAAB01008964 EAA12719.3 GFDL01007600 JAV27445.1 ATLV01014582 KE524975 KFB39089.1 GAMC01015023 JAB91532.1 GAKP01003585 JAC55367.1 GBXI01017000 JAC97291.1 DS233573 EDS30582.1 GDHF01021657 JAI30657.1 OUUW01000010 SPP85569.1 JRES01000300 KNC32610.1 CH379061 EDY70677.2 PYGN01000083 PSN54798.1 AJWK01021695 AJWK01021696 AJWK01021697 AJWK01021698 CH479195 EDW27215.1 GFDF01010958 JAV03126.1 GFDF01010959 JAV03125.1 CH916372 EDV98776.1 CH963857 EDW76215.1 CH933807 EDW12389.2 CH940654 EDW57673.2 SPP85570.1 KK853038 KDR12191.1 CM000362 CM002911 EDX05817.1 KMY91630.1 CP012523 ALC38015.1 NEVH01008240 PNF34346.1 CH954177 EDV59883.2 GBYB01011464 GBYB01011465 GBYB01011466 JAG81231.1 JAG81232.1 JAG81233.1 CCAG010000397 UFQS01000381 UFQT01000381 SSX03421.1 SSX23786.1 AE013599 BT128906 AAM68369.1 AEO72333.1 KQ776753 OAD52186.1 KQ976401 KYM92480.1 AY071311 AAL48933.1 ADTU01013285 KQ414905 KOC59604.1 CH902638 KPU75596.1 AJVK01011049 AJVK01011050 AJVK01011051 KQ978625 KYN29669.1 GL888387 EGI61817.1 KQ977444 KYN02665.1 KK107119 EZA58746.1 KQ435709 KOX80052.1 KZ288312 PBC28410.1 NNAY01003371 OXU19531.1 GL439579 EFN67012.1 GDHF01003457 JAI48857.1 GALA01001496 JAA93356.1 DS232397 EDS41118.1 DS235811 EEB17350.1 GDIQ01202975 GDIP01091067 JAK48750.1 JAM12648.1 GDIQ01256924 JAJ94800.1 GDIP01020372 JAM83343.1 GDIQ01250804 JAK00921.1 GDIP01043218 JAM60497.1 GL732528 EFX87100.1
Proteomes
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
UP000075885
+ More
UP000007062 UP000076407 UP000030765 UP000002320 UP000268350 UP000037069 UP000001819 UP000245037 UP000092461 UP000008744 UP000001070 UP000007798 UP000009192 UP000078200 UP000092445 UP000091820 UP000008792 UP000027135 UP000000304 UP000092553 UP000235965 UP000008711 UP000092444 UP000000803 UP000095301 UP000078540 UP000005205 UP000053825 UP000007801 UP000092462 UP000078492 UP000007755 UP000078542 UP000053097 UP000005203 UP000053105 UP000242457 UP000215335 UP000000311 UP000009046 UP000000305
UP000007062 UP000076407 UP000030765 UP000002320 UP000268350 UP000037069 UP000001819 UP000245037 UP000092461 UP000008744 UP000001070 UP000007798 UP000009192 UP000078200 UP000092445 UP000091820 UP000008792 UP000027135 UP000000304 UP000092553 UP000235965 UP000008711 UP000092444 UP000000803 UP000095301 UP000078540 UP000005205 UP000053825 UP000007801 UP000092462 UP000078492 UP000007755 UP000078542 UP000053097 UP000005203 UP000053105 UP000242457 UP000215335 UP000000311 UP000009046 UP000000305
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1WIW1
A0A2A4K3D3
A0A2W1C137
A0A0L7LSG1
A0A194PKA6
A0A194RCJ2
+ More
A0A212ETW4 A0A1E1W6L8 A0A182PAQ5 Q7Q4K1 A0A1S4GY95 A0A182XFT5 A0A1Q3FIU6 A0A084VM95 W8AZ70 A0A034WLY2 A0A0A1WEJ1 B0XJP3 A0A0K8UVF9 A0A3B0KGT0 A0A0L0CK33 B5DJA8 A0A2P8ZE82 A0A1B0GJH4 B4GXA4 A0A1L8D9G5 A0A1L8D9T6 B4JPE6 B4MUM0 B4KI96 A0A1A9VIT8 A0A1A9Z408 A0A1A9WXH8 B4M8Z7 A0A3B0KBM8 A0A067R345 B4QCM8 A0A0M4E662 A0A2J7R0K1 B3N3K1 A0A0C9RS63 A0A1B0FEZ2 A0A336M0D1 A1Z6G4 A0A310S727 A0A1I8MQZ2 A0A195BW22 Q8SYU4 A0A158NEF7 A0A0L7QM12 A0A0N8NZY2 A0A1B0D3V6 A0A195EP58 F4WVJ6 A0A195CPV8 A0A026WSU4 A0A088A6Q4 A0A0N0U781 A0A2A3E9L5 A0A232EMG5 E2AHT3 A0A0K8WCP7 T1DGX6 B0X5Y7 E0VVE4 A0A0P5J8X8 A0A0P5FBA1 A0A0P6B3Y3 A0A0P5G5E8 A0A0P6AG28 E9FZQ8
A0A212ETW4 A0A1E1W6L8 A0A182PAQ5 Q7Q4K1 A0A1S4GY95 A0A182XFT5 A0A1Q3FIU6 A0A084VM95 W8AZ70 A0A034WLY2 A0A0A1WEJ1 B0XJP3 A0A0K8UVF9 A0A3B0KGT0 A0A0L0CK33 B5DJA8 A0A2P8ZE82 A0A1B0GJH4 B4GXA4 A0A1L8D9G5 A0A1L8D9T6 B4JPE6 B4MUM0 B4KI96 A0A1A9VIT8 A0A1A9Z408 A0A1A9WXH8 B4M8Z7 A0A3B0KBM8 A0A067R345 B4QCM8 A0A0M4E662 A0A2J7R0K1 B3N3K1 A0A0C9RS63 A0A1B0FEZ2 A0A336M0D1 A1Z6G4 A0A310S727 A0A1I8MQZ2 A0A195BW22 Q8SYU4 A0A158NEF7 A0A0L7QM12 A0A0N8NZY2 A0A1B0D3V6 A0A195EP58 F4WVJ6 A0A195CPV8 A0A026WSU4 A0A088A6Q4 A0A0N0U781 A0A2A3E9L5 A0A232EMG5 E2AHT3 A0A0K8WCP7 T1DGX6 B0X5Y7 E0VVE4 A0A0P5J8X8 A0A0P5FBA1 A0A0P6B3Y3 A0A0P5G5E8 A0A0P6AG28 E9FZQ8
Ontologies
PANTHER
Topology
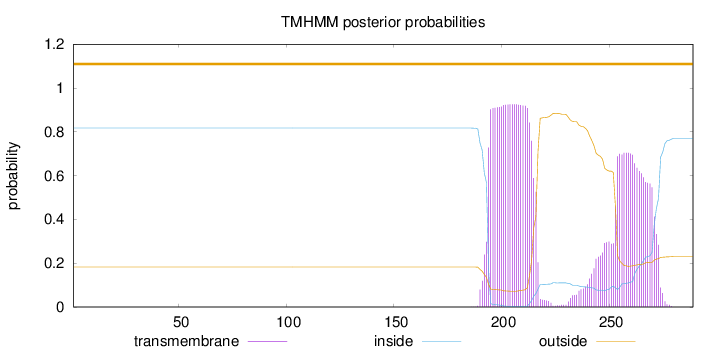
Length:
289
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
37.30586
Exp number, first 60 AAs:
0.00168
Total prob of N-in:
0.81753
outside
1 - 289
Population Genetic Test Statistics
Pi
186.618104
Theta
157.564177
Tajima's D
-0.599563
CLR
0.878881
CSRT
0.218139093045348
Interpretation
Uncertain
