Pre Gene Modal
BGIBMGA005938
Annotation
PREDICTED:_uncharacterized_protein_LOC101739791_isoform_X4_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.683
Sequence
CDS
ATGAGAGTTTTAAGTAATTATGACCTTAGTGGATATACAGCGCTCCAAATTTACTGTAGAGGTCAAGGACAATTTAATCGTTTCAAAGTAACTTTACGGCACAAAGGTTTTAATGATGAAACGAATTATTCTTTCGAACAATATTTTCAGGCTCCCAAAGATGAATTCGCTGTACGTGATTTAAGATTATCAGAATTCAAGGCCTACTACCGAGGGAAAAAAGTAGTTAATAACGAGACTTTGAATTTATCGCAAATTACCAGCATTGGATTGCAAATGTACGGCGGAGTGTATCAAACAGTGAAACAAAAAGGACCTGCAACTTTAGAAATTGATTGGATTAGAGCGGTTGTACTTTAG
Protein
MRVLSNYDLSGYTALQIYCRGQGQFNRFKVTLRHKGFNDETNYSFEQYFQAPKDEFAVRDLRLSEFKAYYRGKKVVNNETLNLSQITSIGLQMYGGVYQTVKQKGPATLEIDWIRAVVL
Summary
Similarity
Belongs to the peptidase S9B family.
Uniprot
H9J8U6
A0A2H1WH36
A0A2A4K3T2
A0A212ETX3
A0A2A4K3F6
A0A2W1BWW4
+ More
S4NZH6 A0A0L7LSI0 A0A194RD39 A0A194PKL9 A9NWT2 A0A1B6MGC9 A0A1B6MGZ6 A0A2R7VXP0 A0A1S3DTK5 A0A1B6JF70 A0A146LBM6 A0A146LYN1 A0A0K8TCY6 A0A1B6CS23 A0A026WJK9 A0A1B6F8Q8 A0A0P5GWS1 A0A0P5ZT23 A0A0P5Z552 A0A0P6E6W0 A0A0P6BVQ3 E9IRE1 A0A0P6E0D5 A0A0P5NDU2 E2BTY1 A0A2A3EJR0 A0A088A3U9 A0A0N0U615 F4W7C2 A0A310SXR9 A0A232FNC6 A0A0C9QJJ0 K7IXE9 A0A0C9RNB8 A0A195DSK0 A0A195EX81 A0A195AU45 A0A158NPW6 A0A154P0A6 A0A2S2NKG7 A0A151X8U1 A0A0P5FUD4 A0A1B6K875 A0A1B6LBG0 C4WV72 A0A0N8DLS0 A0A0P5CTB6 A0A232FN40 K7JDH8 V5GWP3 A0A0X3Q272 A0A183S800 A0A0X3NKA0 A0A094ZWF6 A0A094ZAK8 A0A183P951 A0A183K0I8 A0A183QML2 A0A183QKZ6 A0A3P7LCL8 G4VDQ3 A0A0P5TE13 Q5DAC6 A0A3P7MSV8 A0A3P7L0H2 C1LJG7 A0A139WGZ5 D6WL31 A0A183MLY3 A0A3P8FH79 A0A183NQR6 G4VDQ0 A0A183MLY5 A0A3P8CT25 C7TZP2 A0A158QBI8 A0A1Z9CRJ1 A0A0R3T3H6 A0A183LI01 A0A316TUY0 A0A0R3W5P5 A0A183LI00 A0A183Q270 A0A0S4T8Y2 A0A0S8FBX4 A0A1Z9PZ71 A0A2D9TM29 A0A183NLN5 A0A2E9TGE4 A0A2N0VLP3
S4NZH6 A0A0L7LSI0 A0A194RD39 A0A194PKL9 A9NWT2 A0A1B6MGC9 A0A1B6MGZ6 A0A2R7VXP0 A0A1S3DTK5 A0A1B6JF70 A0A146LBM6 A0A146LYN1 A0A0K8TCY6 A0A1B6CS23 A0A026WJK9 A0A1B6F8Q8 A0A0P5GWS1 A0A0P5ZT23 A0A0P5Z552 A0A0P6E6W0 A0A0P6BVQ3 E9IRE1 A0A0P6E0D5 A0A0P5NDU2 E2BTY1 A0A2A3EJR0 A0A088A3U9 A0A0N0U615 F4W7C2 A0A310SXR9 A0A232FNC6 A0A0C9QJJ0 K7IXE9 A0A0C9RNB8 A0A195DSK0 A0A195EX81 A0A195AU45 A0A158NPW6 A0A154P0A6 A0A2S2NKG7 A0A151X8U1 A0A0P5FUD4 A0A1B6K875 A0A1B6LBG0 C4WV72 A0A0N8DLS0 A0A0P5CTB6 A0A232FN40 K7JDH8 V5GWP3 A0A0X3Q272 A0A183S800 A0A0X3NKA0 A0A094ZWF6 A0A094ZAK8 A0A183P951 A0A183K0I8 A0A183QML2 A0A183QKZ6 A0A3P7LCL8 G4VDQ3 A0A0P5TE13 Q5DAC6 A0A3P7MSV8 A0A3P7L0H2 C1LJG7 A0A139WGZ5 D6WL31 A0A183MLY3 A0A3P8FH79 A0A183NQR6 G4VDQ0 A0A183MLY5 A0A3P8CT25 C7TZP2 A0A158QBI8 A0A1Z9CRJ1 A0A0R3T3H6 A0A183LI01 A0A316TUY0 A0A0R3W5P5 A0A183LI00 A0A183Q270 A0A0S4T8Y2 A0A0S8FBX4 A0A1Z9PZ71 A0A2D9TM29 A0A183NLN5 A0A2E9TGE4 A0A2N0VLP3
Pubmed
EMBL
BABH01005042
ODYU01008632
SOQ52383.1
NWSH01000193
PCG78558.1
AGBW02012530
+ More
OWR44929.1 PCG78556.1 KZ149895 PZC78831.1 GAIX01013385 JAA79175.1 JTDY01000181 KOB78412.1 KQ460398 KPJ15190.1 KQ459601 KPI93872.1 EF085797 ABK25093.1 GEBQ01004967 JAT35010.1 GEBQ01004812 JAT35165.1 KK854110 PTY11450.1 GECU01009918 JAS97788.1 GDHC01014127 JAQ04502.1 GDHC01006883 JAQ11746.1 GBRD01002394 GBRD01002393 GDHC01002212 JAG63428.1 JAQ16417.1 GEDC01020972 JAS16326.1 KK107183 QOIP01000011 EZA55861.1 RLU16656.1 GECZ01028879 GECZ01023147 GECZ01018908 GECZ01015986 JAS40890.1 JAS46622.1 JAS50861.1 JAS53783.1 GDIQ01241514 JAK10211.1 GDIP01043153 LRGB01001348 JAM60562.1 KZS12906.1 GDIP01049075 JAM54640.1 GDIQ01066777 JAN27960.1 GDIP01009100 JAM94615.1 GL765107 EFZ16864.1 GDIQ01084621 JAN10116.1 GDIQ01150247 JAL01479.1 GL450531 EFN80880.1 KZ288225 PBC31930.1 KQ435759 KOX75732.1 GL887844 EGI69795.1 KQ759768 OAD62949.1 NNAY01000020 OXU31877.1 GBYB01014988 JAG84755.1 AAZX01000007 GBYB01014987 JAG84754.1 KQ980487 KYN15895.1 KQ981931 KYN32833.1 KQ976745 KYM75504.1 ADTU01022791 KQ434787 KZC05297.1 GGMR01005055 MBY17674.1 KQ982409 KYQ56749.1 GDIQ01255933 JAJ95791.1 GECU01000075 JAT07632.1 GEBQ01018945 JAT21032.1 ABLF02018531 AK341386 BAH71792.1 GDIP01021287 JAM82428.1 GDIP01169896 JAJ53506.1 OXU31878.1 GALX01003788 JAB64678.1 GEEE01004912 JAP58313.1 UYSU01000230 VDL85458.1 GEEE01022831 JAP40394.1 KL251054 KGB38657.1 AMPZ01023331 KGB31275.1 UZAL01030967 VDP56341.1 UZAK01032772 VDP31090.1 UYRU01060407 VDN14795.1 HE601625 CCD76990.1 GDIP01129211 JAL74503.1 AY815498 AAW27230.1 UYRU01071661 VDN21281.1 UYRU01050345 VDN10955.1 FN319115 FN319116 FN319117 CAX74845.1 KQ971343 KYB27169.1 EFA03518.2 UZAI01017278 VDP22829.1 UZAL01012738 VDP07164.1 CCD76987.1 VDP22834.1 FN330859 CAX83076.1 UYSG01000006 VDL11686.1 UZAE01000620 VDN97418.1 UZAI01000975 VDO58040.1 QGGB01000005 PWN06895.1 UYRS01018416 VDK35131.1 VDO58039.1 UZAL01045170 VDP83195.1 LJTI01000087 KPK57197.1 NZUF01000005 MAQ21734.1 UZAL01004845 VDO91293.1 PCAL01000008 MBU82291.1 PISP01000001 PKD45102.1
OWR44929.1 PCG78556.1 KZ149895 PZC78831.1 GAIX01013385 JAA79175.1 JTDY01000181 KOB78412.1 KQ460398 KPJ15190.1 KQ459601 KPI93872.1 EF085797 ABK25093.1 GEBQ01004967 JAT35010.1 GEBQ01004812 JAT35165.1 KK854110 PTY11450.1 GECU01009918 JAS97788.1 GDHC01014127 JAQ04502.1 GDHC01006883 JAQ11746.1 GBRD01002394 GBRD01002393 GDHC01002212 JAG63428.1 JAQ16417.1 GEDC01020972 JAS16326.1 KK107183 QOIP01000011 EZA55861.1 RLU16656.1 GECZ01028879 GECZ01023147 GECZ01018908 GECZ01015986 JAS40890.1 JAS46622.1 JAS50861.1 JAS53783.1 GDIQ01241514 JAK10211.1 GDIP01043153 LRGB01001348 JAM60562.1 KZS12906.1 GDIP01049075 JAM54640.1 GDIQ01066777 JAN27960.1 GDIP01009100 JAM94615.1 GL765107 EFZ16864.1 GDIQ01084621 JAN10116.1 GDIQ01150247 JAL01479.1 GL450531 EFN80880.1 KZ288225 PBC31930.1 KQ435759 KOX75732.1 GL887844 EGI69795.1 KQ759768 OAD62949.1 NNAY01000020 OXU31877.1 GBYB01014988 JAG84755.1 AAZX01000007 GBYB01014987 JAG84754.1 KQ980487 KYN15895.1 KQ981931 KYN32833.1 KQ976745 KYM75504.1 ADTU01022791 KQ434787 KZC05297.1 GGMR01005055 MBY17674.1 KQ982409 KYQ56749.1 GDIQ01255933 JAJ95791.1 GECU01000075 JAT07632.1 GEBQ01018945 JAT21032.1 ABLF02018531 AK341386 BAH71792.1 GDIP01021287 JAM82428.1 GDIP01169896 JAJ53506.1 OXU31878.1 GALX01003788 JAB64678.1 GEEE01004912 JAP58313.1 UYSU01000230 VDL85458.1 GEEE01022831 JAP40394.1 KL251054 KGB38657.1 AMPZ01023331 KGB31275.1 UZAL01030967 VDP56341.1 UZAK01032772 VDP31090.1 UYRU01060407 VDN14795.1 HE601625 CCD76990.1 GDIP01129211 JAL74503.1 AY815498 AAW27230.1 UYRU01071661 VDN21281.1 UYRU01050345 VDN10955.1 FN319115 FN319116 FN319117 CAX74845.1 KQ971343 KYB27169.1 EFA03518.2 UZAI01017278 VDP22829.1 UZAL01012738 VDP07164.1 CCD76987.1 VDP22834.1 FN330859 CAX83076.1 UYSG01000006 VDL11686.1 UZAE01000620 VDN97418.1 UZAI01000975 VDO58040.1 QGGB01000005 PWN06895.1 UYRS01018416 VDK35131.1 VDO58039.1 UZAL01045170 VDP83195.1 LJTI01000087 KPK57197.1 NZUF01000005 MAQ21734.1 UZAL01004845 VDO91293.1 PCAL01000008 MBU82291.1 PISP01000001 PKD45102.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
+ More
UP000079169 UP000053097 UP000279307 UP000076858 UP000008237 UP000242457 UP000005203 UP000053105 UP000007755 UP000215335 UP000002358 UP000078492 UP000078541 UP000078540 UP000005205 UP000076502 UP000075809 UP000007819 UP000050788 UP000275846 UP000050791 UP000050789 UP000050792 UP000008854 UP000007266 UP000050790 UP000277204 UP000046397 UP000274504 UP000046398 UP000278807 UP000245533 UP000046400 UP000282613 UP000051321 UP000233398
UP000079169 UP000053097 UP000279307 UP000076858 UP000008237 UP000242457 UP000005203 UP000053105 UP000007755 UP000215335 UP000002358 UP000078492 UP000078541 UP000078540 UP000005205 UP000076502 UP000075809 UP000007819 UP000050788 UP000275846 UP000050791 UP000050789 UP000050792 UP000008854 UP000007266 UP000050790 UP000277204 UP000046397 UP000274504 UP000046398 UP000278807 UP000245533 UP000046400 UP000282613 UP000051321 UP000233398
PRIDE
Pfam
Interpro
IPR039131
NDUFAF1
+ More
IPR013857 NADH-UbQ_OxRdtase-assoc_prot30
IPR008979 Galactose-bd-like_sf
IPR005225 Small_GTP-bd_dom
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR030702 Rab14
IPR013578 Peptidase_M16C_assoc
IPR002469 Peptidase_S9B_N
IPR011249 Metalloenz_LuxS/M16
IPR001375 Peptidase_S9
IPR011765 Pept_M16_N
IPR038554 Peptidase_S9B_N_sf
IPR029058 AB_hydrolase
IPR007863 Peptidase_M16_C
IPR013857 NADH-UbQ_OxRdtase-assoc_prot30
IPR008979 Galactose-bd-like_sf
IPR005225 Small_GTP-bd_dom
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR030702 Rab14
IPR013578 Peptidase_M16C_assoc
IPR002469 Peptidase_S9B_N
IPR011249 Metalloenz_LuxS/M16
IPR001375 Peptidase_S9
IPR011765 Pept_M16_N
IPR038554 Peptidase_S9B_N_sf
IPR029058 AB_hydrolase
IPR007863 Peptidase_M16_C
Gene 3D
CDD
ProteinModelPortal
H9J8U6
A0A2H1WH36
A0A2A4K3T2
A0A212ETX3
A0A2A4K3F6
A0A2W1BWW4
+ More
S4NZH6 A0A0L7LSI0 A0A194RD39 A0A194PKL9 A9NWT2 A0A1B6MGC9 A0A1B6MGZ6 A0A2R7VXP0 A0A1S3DTK5 A0A1B6JF70 A0A146LBM6 A0A146LYN1 A0A0K8TCY6 A0A1B6CS23 A0A026WJK9 A0A1B6F8Q8 A0A0P5GWS1 A0A0P5ZT23 A0A0P5Z552 A0A0P6E6W0 A0A0P6BVQ3 E9IRE1 A0A0P6E0D5 A0A0P5NDU2 E2BTY1 A0A2A3EJR0 A0A088A3U9 A0A0N0U615 F4W7C2 A0A310SXR9 A0A232FNC6 A0A0C9QJJ0 K7IXE9 A0A0C9RNB8 A0A195DSK0 A0A195EX81 A0A195AU45 A0A158NPW6 A0A154P0A6 A0A2S2NKG7 A0A151X8U1 A0A0P5FUD4 A0A1B6K875 A0A1B6LBG0 C4WV72 A0A0N8DLS0 A0A0P5CTB6 A0A232FN40 K7JDH8 V5GWP3 A0A0X3Q272 A0A183S800 A0A0X3NKA0 A0A094ZWF6 A0A094ZAK8 A0A183P951 A0A183K0I8 A0A183QML2 A0A183QKZ6 A0A3P7LCL8 G4VDQ3 A0A0P5TE13 Q5DAC6 A0A3P7MSV8 A0A3P7L0H2 C1LJG7 A0A139WGZ5 D6WL31 A0A183MLY3 A0A3P8FH79 A0A183NQR6 G4VDQ0 A0A183MLY5 A0A3P8CT25 C7TZP2 A0A158QBI8 A0A1Z9CRJ1 A0A0R3T3H6 A0A183LI01 A0A316TUY0 A0A0R3W5P5 A0A183LI00 A0A183Q270 A0A0S4T8Y2 A0A0S8FBX4 A0A1Z9PZ71 A0A2D9TM29 A0A183NLN5 A0A2E9TGE4 A0A2N0VLP3
S4NZH6 A0A0L7LSI0 A0A194RD39 A0A194PKL9 A9NWT2 A0A1B6MGC9 A0A1B6MGZ6 A0A2R7VXP0 A0A1S3DTK5 A0A1B6JF70 A0A146LBM6 A0A146LYN1 A0A0K8TCY6 A0A1B6CS23 A0A026WJK9 A0A1B6F8Q8 A0A0P5GWS1 A0A0P5ZT23 A0A0P5Z552 A0A0P6E6W0 A0A0P6BVQ3 E9IRE1 A0A0P6E0D5 A0A0P5NDU2 E2BTY1 A0A2A3EJR0 A0A088A3U9 A0A0N0U615 F4W7C2 A0A310SXR9 A0A232FNC6 A0A0C9QJJ0 K7IXE9 A0A0C9RNB8 A0A195DSK0 A0A195EX81 A0A195AU45 A0A158NPW6 A0A154P0A6 A0A2S2NKG7 A0A151X8U1 A0A0P5FUD4 A0A1B6K875 A0A1B6LBG0 C4WV72 A0A0N8DLS0 A0A0P5CTB6 A0A232FN40 K7JDH8 V5GWP3 A0A0X3Q272 A0A183S800 A0A0X3NKA0 A0A094ZWF6 A0A094ZAK8 A0A183P951 A0A183K0I8 A0A183QML2 A0A183QKZ6 A0A3P7LCL8 G4VDQ3 A0A0P5TE13 Q5DAC6 A0A3P7MSV8 A0A3P7L0H2 C1LJG7 A0A139WGZ5 D6WL31 A0A183MLY3 A0A3P8FH79 A0A183NQR6 G4VDQ0 A0A183MLY5 A0A3P8CT25 C7TZP2 A0A158QBI8 A0A1Z9CRJ1 A0A0R3T3H6 A0A183LI01 A0A316TUY0 A0A0R3W5P5 A0A183LI00 A0A183Q270 A0A0S4T8Y2 A0A0S8FBX4 A0A1Z9PZ71 A0A2D9TM29 A0A183NLN5 A0A2E9TGE4 A0A2N0VLP3
Ontologies
GO
PANTHER
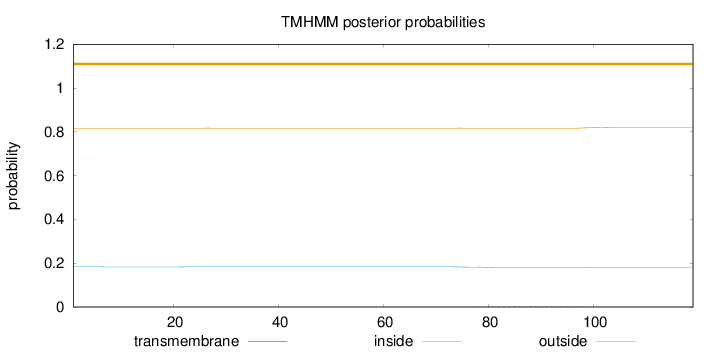
Topology
Length:
119
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0958
Exp number, first 60 AAs:
0.00175
Total prob of N-in:
0.18330
outside
1 - 119
Population Genetic Test Statistics
Pi
211.848011
Theta
172.567374
Tajima's D
0.755555
CLR
0.002411
CSRT
0.589020548972551
Interpretation
Uncertain
