Pre Gene Modal
BGIBMGA006208
Annotation
PREDICTED:_RNA-binding_protein_39_[Papilio_polytes]
Location in the cell
Nuclear Reliability : 1.292
Sequence
CDS
ATGGCGGAAGATTTTGATGTTGAGGCAATGTTAGAAGCTCCATATAATAAATCGGATAACACTTCCTCATCAAAGCATCGATCGGACAAGCACTCAGATAAGCACAAAGAAAGGGATAAAGATGGTTCAAGAAGGCGTAGTCGATCGAGAGATAAAAGAAGATCAAGGGATAAGGATTCACGTCGTTCAAGAGATCGAGACGATAAAAAAGACAGAGAAAGAGGCAGAGACAGAGATCGTCGAGATCGCGACAAAGATCGGATTAAGGATAAAGACCGCGAGAGAGAAAGAGAACGAGATAAAGAAAAACGTCGTAGCCGCGACAAGGACCGAAGTCGGGAAAGAGATCGCAGTCGCGATAAAAACGTTCATAAAAAAGAAAAAACTCCTGAAAGAGAACCACAAAAAGAATATAGGTCAAAGTCGAGGGGGCTAGACCCGAAACTTGAAGATCTACCTCCAGAAGAAAGAGATCTAAGAACTGTATTTTGCATGCAATTGTCGCAAAGAATCCGTGCTAAAGATTTAGAAGAATTTTTCTCATCAGTTGGCAAGGTTCGAGACGTAAGACTTATTACTTGCAATAAAACAAGAAGATTCAAAGGTATTGCATATATTGAATTCAAAGATGCTGAATCAGTGCCTTTGGCTCTTGGCTTAACAGGACAAAAGTTACTTGGAGTTCCTATAATTGTACAGCACACGCAAGCCGAAAAAAATAGGGTTGGCAATACATTGCCGAATTTAGCACCAAAGACCAGTAATGGACCTACTAGACTCTATGTTGGGTCTTTACATTTTAATATTACAGAGGATATGCTACGTGGCATTTTTGAACCATTCGGAAAAATTGATCACATTCAACTAATGACTGATCCAGAAACTGGTAAAAGTAAAGGATATGGATTTCTCACTTTCCACCATGCCAGTGATGCTAAGAAGGCTATGGAACAGTTAAATGGTTTTGAGTTAGCTGGGCGGCCTATGAAAGTTGGAAACGTCACAGAACGTGCAGATGGAGGATCAAGCACGCGGTTTGATGCTGATGAGTTGGATCGAGCTGGAATTGACCTTGGTGCCACGGGCCGTTTACAATTAATGTTTAAATTAGCAGAGGGAACAGGCTTACAAATACCTCCTGCAGCAGCATCAGTACTAATGGGGAGTGGTTCAGCTTTAGTGTCTCCGCAACCTCAAGTTGCACCTCCTATTGCAACACAATGCTTCATGCTCAATAACATGTTTGATCCAGCAACGGAAACAAATCCAAGTTGGGATATTGAAATAAGGGATGATGTTATCAGTGAATGCAACAAACACGGTGGAGTGTTGCATGTGTATGTTGATAAAGCATCGCCTCAGGGAAATGTGTACTGCAAGTGTCCGACAATAGCGACGGCCGTTGCCTCCGTGAACAGTTTGCACGGAAGATGGTTTGCGGGTCGTGTTATTACGGCGGCGTATGTACCGCTCGTCAATTATCATTCTTTATTCCCGGACGCGATGACTGCACTCAGCTTACTATTGCCTTCGAGGCAGCGATAG
Protein
MAEDFDVEAMLEAPYNKSDNTSSSKHRSDKHSDKHKERDKDGSRRRSRSRDKRRSRDKDSRRSRDRDDKKDRERGRDRDRRDRDKDRIKDKDRERERERDKEKRRSRDKDRSRERDRSRDKNVHKKEKTPEREPQKEYRSKSRGLDPKLEDLPPEERDLRTVFCMQLSQRIRAKDLEEFFSSVGKVRDVRLITCNKTRRFKGIAYIEFKDAESVPLALGLTGQKLLGVPIIVQHTQAEKNRVGNTLPNLAPKTSNGPTRLYVGSLHFNITEDMLRGIFEPFGKIDHIQLMTDPETGKSKGYGFLTFHHASDAKKAMEQLNGFELAGRPMKVGNVTERADGGSSTRFDADELDRAGIDLGATGRLQLMFKLAEGTGLQIPPAAASVLMGSGSALVSPQPQVAPPIATQCFMLNNMFDPATETNPSWDIEIRDDVISECNKHGGVLHVYVDKASPQGNVYCKCPTIATAVASVNSLHGRWFAGRVITAAYVPLVNYHSLFPDAMTALSLLLPSRQR
Summary
Uniprot
H9J9L5
A0A2A4K3S6
A0A2W1BUS7
A0A2H1WHL4
A0A194PLJ8
A0A194RGY3
+ More
A0A212ETV9 A0A0L7LSI4 A0A2J7R0F3 A0A0K8T0Y2 A0A0K8T8Z7 A0A1W4X3F2 A0A0N7Z915 T1HXL9 A0A1S4EFA0 A0A3Q0IZ48 N6U3V4 D6WLZ9 A0A0T6BB65 A0A069DU20 A0A1Y1M1J5 A0A1Y1LZK6 A0A154PRG0 A0A0J7KN76 A0A310SIX9 E2C448 E2ACV0 A0A026WGX8 A0A195CUK8 A0A151JP25 F4X152 A0A195BZ42 A0A195F0G1 A0A158P3P2 A0A087ZVF7 A0A0C9QWU2 A0A151XG88 A0A0N0BKB9 A0A0C9RGZ9 A0A1B6MNS3 A0A1B6LBP5 E0VR69 A0A0L0C7T0 A0A0K8TLP1 A0A0K8VQV4 A0A034WIX8 A0A1A9WXG2 A0A0A1WXC4 A0A1L8DFE9 A0A1B0FEY8 T1PGA1 A0A1A9Z413 W8BUP4 A0A1B0BVM3 A0A0L7QS78 A0A1A9XMK5 B0X5Y4 A0A182QYA1 A0A1Q3F788 A0A1Q3F7L1 A0A1Q3F797 B0XJP6 A0A1Q3F7F0 Q17B91 A0A084VM93 A0A182VJI7 A0A182JBV7 A0A182L0E0 A0A182RRQ0 A0A1S4GYY7 A0A182TN11 A0A2C9GNI9 A0A182NUV4 U5EWK0 Q7PN29 A0A182KCI9 A0A182XFT4 A0A023ETK7 A0A2S2QPG9 J9JQ40 A0A336LJK2 A0A182WHD7 A0A182PAQ6 A0A182IC17 A0A2S2P5L6 A0A1I8PK21 A0A1I8PJW5 A0A1I8PJT8 A0A182NES8 A0A182Y9Z6 B4JPE9
A0A212ETV9 A0A0L7LSI4 A0A2J7R0F3 A0A0K8T0Y2 A0A0K8T8Z7 A0A1W4X3F2 A0A0N7Z915 T1HXL9 A0A1S4EFA0 A0A3Q0IZ48 N6U3V4 D6WLZ9 A0A0T6BB65 A0A069DU20 A0A1Y1M1J5 A0A1Y1LZK6 A0A154PRG0 A0A0J7KN76 A0A310SIX9 E2C448 E2ACV0 A0A026WGX8 A0A195CUK8 A0A151JP25 F4X152 A0A195BZ42 A0A195F0G1 A0A158P3P2 A0A087ZVF7 A0A0C9QWU2 A0A151XG88 A0A0N0BKB9 A0A0C9RGZ9 A0A1B6MNS3 A0A1B6LBP5 E0VR69 A0A0L0C7T0 A0A0K8TLP1 A0A0K8VQV4 A0A034WIX8 A0A1A9WXG2 A0A0A1WXC4 A0A1L8DFE9 A0A1B0FEY8 T1PGA1 A0A1A9Z413 W8BUP4 A0A1B0BVM3 A0A0L7QS78 A0A1A9XMK5 B0X5Y4 A0A182QYA1 A0A1Q3F788 A0A1Q3F7L1 A0A1Q3F797 B0XJP6 A0A1Q3F7F0 Q17B91 A0A084VM93 A0A182VJI7 A0A182JBV7 A0A182L0E0 A0A182RRQ0 A0A1S4GYY7 A0A182TN11 A0A2C9GNI9 A0A182NUV4 U5EWK0 Q7PN29 A0A182KCI9 A0A182XFT4 A0A023ETK7 A0A2S2QPG9 J9JQ40 A0A336LJK2 A0A182WHD7 A0A182PAQ6 A0A182IC17 A0A2S2P5L6 A0A1I8PK21 A0A1I8PJW5 A0A1I8PJT8 A0A182NES8 A0A182Y9Z6 B4JPE9
Pubmed
EMBL
BABH01005042
NWSH01000193
PCG78554.1
KZ149895
PZC78832.1
ODYU01008632
+ More
SOQ52382.1 KQ459601 KPI93873.1 KQ460398 KPJ15191.1 AGBW02012530 OWR44930.1 JTDY01000181 KOB78417.1 NEVH01008240 PNF34319.1 GBRD01006624 JAG59197.1 GBRD01003925 JAG61896.1 GDKW01001811 JAI54784.1 ACPB03010247 ACPB03010248 APGK01040756 KB740984 KB631904 ENN76275.1 ERL87102.1 KQ971343 EFA04195.1 LJIG01002607 KRT84333.1 GBGD01001281 JAC87608.1 GEZM01044078 JAV78520.1 GEZM01044077 JAV78521.1 KQ435007 KZC13700.1 LBMM01005229 KMQ91686.1 KQ762207 OAD56040.1 GL452396 EFN77277.1 GL438582 EFN68748.1 KK107250 QOIP01000002 EZA54334.1 RLU25526.1 KQ977304 KYN03824.1 KQ978781 KYN28486.1 GL888526 EGI59819.1 KQ976394 KYM93196.1 KQ981880 KYN33963.1 ADTU01008308 GBYB01005142 JAG74909.1 KQ982174 KYQ59367.1 KQ435701 KOX80381.1 GBYB01007595 JAG77362.1 GEBQ01002425 JAT37552.1 GEBQ01018872 JAT21105.1 DS235459 EEB15875.1 JRES01000794 KNC28312.1 GDAI01002326 JAI15277.1 GDHF01016259 GDHF01011053 JAI36055.1 JAI41261.1 GAKP01005249 JAC53703.1 GBXI01014378 GBXI01011122 JAC99913.1 JAD03170.1 GFDF01008913 JAV05171.1 CCAG010000397 KA647791 AFP62420.1 GAMC01005892 GAMC01005890 JAC00664.1 JXJN01021375 KQ414766 KOC61394.1 DS232397 EDS41115.1 AXCN02000840 GFDL01011614 JAV23431.1 GFDL01011499 JAV23546.1 GFDL01011613 JAV23432.1 DS233573 EDS30585.1 GFDL01011555 JAV23490.1 CH477324 EAT43555.1 ATLV01014582 KE524975 KFB39087.1 AAAB01008964 APCN01000772 GANO01001381 JAB58490.1 EAA12873.4 GAPW01000945 JAC12653.1 GGMS01010217 MBY79420.1 ABLF02036759 UFQT01000016 SSX17815.1 GGMR01012162 MBY24781.1 CH916372 EDV98779.1
SOQ52382.1 KQ459601 KPI93873.1 KQ460398 KPJ15191.1 AGBW02012530 OWR44930.1 JTDY01000181 KOB78417.1 NEVH01008240 PNF34319.1 GBRD01006624 JAG59197.1 GBRD01003925 JAG61896.1 GDKW01001811 JAI54784.1 ACPB03010247 ACPB03010248 APGK01040756 KB740984 KB631904 ENN76275.1 ERL87102.1 KQ971343 EFA04195.1 LJIG01002607 KRT84333.1 GBGD01001281 JAC87608.1 GEZM01044078 JAV78520.1 GEZM01044077 JAV78521.1 KQ435007 KZC13700.1 LBMM01005229 KMQ91686.1 KQ762207 OAD56040.1 GL452396 EFN77277.1 GL438582 EFN68748.1 KK107250 QOIP01000002 EZA54334.1 RLU25526.1 KQ977304 KYN03824.1 KQ978781 KYN28486.1 GL888526 EGI59819.1 KQ976394 KYM93196.1 KQ981880 KYN33963.1 ADTU01008308 GBYB01005142 JAG74909.1 KQ982174 KYQ59367.1 KQ435701 KOX80381.1 GBYB01007595 JAG77362.1 GEBQ01002425 JAT37552.1 GEBQ01018872 JAT21105.1 DS235459 EEB15875.1 JRES01000794 KNC28312.1 GDAI01002326 JAI15277.1 GDHF01016259 GDHF01011053 JAI36055.1 JAI41261.1 GAKP01005249 JAC53703.1 GBXI01014378 GBXI01011122 JAC99913.1 JAD03170.1 GFDF01008913 JAV05171.1 CCAG010000397 KA647791 AFP62420.1 GAMC01005892 GAMC01005890 JAC00664.1 JXJN01021375 KQ414766 KOC61394.1 DS232397 EDS41115.1 AXCN02000840 GFDL01011614 JAV23431.1 GFDL01011499 JAV23546.1 GFDL01011613 JAV23432.1 DS233573 EDS30585.1 GFDL01011555 JAV23490.1 CH477324 EAT43555.1 ATLV01014582 KE524975 KFB39087.1 AAAB01008964 APCN01000772 GANO01001381 JAB58490.1 EAA12873.4 GAPW01000945 JAC12653.1 GGMS01010217 MBY79420.1 ABLF02036759 UFQT01000016 SSX17815.1 GGMR01012162 MBY24781.1 CH916372 EDV98779.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000037510
+ More
UP000235965 UP000192223 UP000015103 UP000079169 UP000019118 UP000030742 UP000007266 UP000076502 UP000036403 UP000008237 UP000000311 UP000053097 UP000279307 UP000078542 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000005203 UP000075809 UP000053105 UP000009046 UP000037069 UP000091820 UP000092444 UP000095301 UP000092445 UP000092460 UP000053825 UP000092443 UP000002320 UP000075886 UP000008820 UP000030765 UP000075903 UP000075880 UP000075882 UP000075900 UP000075902 UP000075840 UP000075884 UP000007062 UP000075881 UP000076407 UP000007819 UP000075920 UP000075885 UP000095300 UP000076408 UP000001070
UP000235965 UP000192223 UP000015103 UP000079169 UP000019118 UP000030742 UP000007266 UP000076502 UP000036403 UP000008237 UP000000311 UP000053097 UP000279307 UP000078542 UP000078492 UP000007755 UP000078540 UP000078541 UP000005205 UP000005203 UP000075809 UP000053105 UP000009046 UP000037069 UP000091820 UP000092444 UP000095301 UP000092445 UP000092460 UP000053825 UP000092443 UP000002320 UP000075886 UP000008820 UP000030765 UP000075903 UP000075880 UP000075882 UP000075900 UP000075902 UP000075840 UP000075884 UP000007062 UP000075881 UP000076407 UP000007819 UP000075920 UP000075885 UP000095300 UP000076408 UP000001070
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9J9L5
A0A2A4K3S6
A0A2W1BUS7
A0A2H1WHL4
A0A194PLJ8
A0A194RGY3
+ More
A0A212ETV9 A0A0L7LSI4 A0A2J7R0F3 A0A0K8T0Y2 A0A0K8T8Z7 A0A1W4X3F2 A0A0N7Z915 T1HXL9 A0A1S4EFA0 A0A3Q0IZ48 N6U3V4 D6WLZ9 A0A0T6BB65 A0A069DU20 A0A1Y1M1J5 A0A1Y1LZK6 A0A154PRG0 A0A0J7KN76 A0A310SIX9 E2C448 E2ACV0 A0A026WGX8 A0A195CUK8 A0A151JP25 F4X152 A0A195BZ42 A0A195F0G1 A0A158P3P2 A0A087ZVF7 A0A0C9QWU2 A0A151XG88 A0A0N0BKB9 A0A0C9RGZ9 A0A1B6MNS3 A0A1B6LBP5 E0VR69 A0A0L0C7T0 A0A0K8TLP1 A0A0K8VQV4 A0A034WIX8 A0A1A9WXG2 A0A0A1WXC4 A0A1L8DFE9 A0A1B0FEY8 T1PGA1 A0A1A9Z413 W8BUP4 A0A1B0BVM3 A0A0L7QS78 A0A1A9XMK5 B0X5Y4 A0A182QYA1 A0A1Q3F788 A0A1Q3F7L1 A0A1Q3F797 B0XJP6 A0A1Q3F7F0 Q17B91 A0A084VM93 A0A182VJI7 A0A182JBV7 A0A182L0E0 A0A182RRQ0 A0A1S4GYY7 A0A182TN11 A0A2C9GNI9 A0A182NUV4 U5EWK0 Q7PN29 A0A182KCI9 A0A182XFT4 A0A023ETK7 A0A2S2QPG9 J9JQ40 A0A336LJK2 A0A182WHD7 A0A182PAQ6 A0A182IC17 A0A2S2P5L6 A0A1I8PK21 A0A1I8PJW5 A0A1I8PJT8 A0A182NES8 A0A182Y9Z6 B4JPE9
A0A212ETV9 A0A0L7LSI4 A0A2J7R0F3 A0A0K8T0Y2 A0A0K8T8Z7 A0A1W4X3F2 A0A0N7Z915 T1HXL9 A0A1S4EFA0 A0A3Q0IZ48 N6U3V4 D6WLZ9 A0A0T6BB65 A0A069DU20 A0A1Y1M1J5 A0A1Y1LZK6 A0A154PRG0 A0A0J7KN76 A0A310SIX9 E2C448 E2ACV0 A0A026WGX8 A0A195CUK8 A0A151JP25 F4X152 A0A195BZ42 A0A195F0G1 A0A158P3P2 A0A087ZVF7 A0A0C9QWU2 A0A151XG88 A0A0N0BKB9 A0A0C9RGZ9 A0A1B6MNS3 A0A1B6LBP5 E0VR69 A0A0L0C7T0 A0A0K8TLP1 A0A0K8VQV4 A0A034WIX8 A0A1A9WXG2 A0A0A1WXC4 A0A1L8DFE9 A0A1B0FEY8 T1PGA1 A0A1A9Z413 W8BUP4 A0A1B0BVM3 A0A0L7QS78 A0A1A9XMK5 B0X5Y4 A0A182QYA1 A0A1Q3F788 A0A1Q3F7L1 A0A1Q3F797 B0XJP6 A0A1Q3F7F0 Q17B91 A0A084VM93 A0A182VJI7 A0A182JBV7 A0A182L0E0 A0A182RRQ0 A0A1S4GYY7 A0A182TN11 A0A2C9GNI9 A0A182NUV4 U5EWK0 Q7PN29 A0A182KCI9 A0A182XFT4 A0A023ETK7 A0A2S2QPG9 J9JQ40 A0A336LJK2 A0A182WHD7 A0A182PAQ6 A0A182IC17 A0A2S2P5L6 A0A1I8PK21 A0A1I8PJW5 A0A1I8PJT8 A0A182NES8 A0A182Y9Z6 B4JPE9
PDB
5CXT
E-value=7.16659e-44,
Score=447
Ontologies
GO
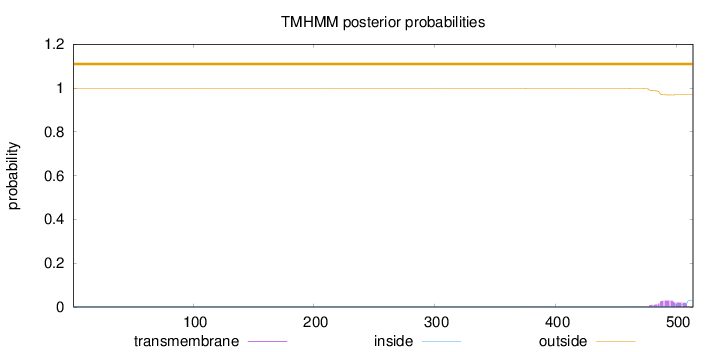
Topology
Length:
514
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.66834
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00186
outside
1 - 514
Population Genetic Test Statistics
Pi
271.045846
Theta
224.787113
Tajima's D
0.665667
CLR
0.081938
CSRT
0.567171641417929
Interpretation
Uncertain
