Gene
KWMTBOMO01912
Pre Gene Modal
BGIBMGA005940
Annotation
Glass_bottom_boat_[Operophtera_brumata]
Full name
ATP-dependent (S)-NAD(P)H-hydrate dehydratase
+ More
Protein 60A
Protein 60A
Alternative Name
ATP-dependent NAD(P)HX dehydratase
Protein glass bottom boat
Protein glass bottom boat
Location in the cell
Nuclear Reliability : 1.921
Sequence
CDS
ATGGCTGGTTTTGTGCAGTTGGTGTGTGCGTGTTTTGCATTGTTTACTGTGGTCACCGATGCCATACTTTCCGGACTTTACATTGACAATGGTGTTGATCAGACGATAATTCATCATACGATTACACGACACGAGCGCATGGTAGTAGAACATGAAATTTTAGATCTTTTAGGCTTAGGAGAAAGACCCAGAAGATCTAAATCTCCACCTTTGGATCGTTCGGCTCCAAGTTTCCTGTTGGACGTTTATAAGCAACTGGCTGAGGAGCACGAACAGGCACGCCCTACACGTAGTTCTGACATGGCTCTCAGCGGTGATGAGCAGCAAGCTATTGATGAAAGTGATTTAATTATGACTTTTCAAAGCAAAAGTAAGTTATTTTTTGTTTTCATTGCTATTTTATGTTAA
Protein
MAGFVQLVCACFALFTVVTDAILSGLYIDNGVDQTIIHHTITRHERMVVEHEILDLLGLGERPRRSKSPPLDRSAPSFLLDVYKQLAEEHEQARPTRSSDMALSGDEQQAIDESDLIMTFQSKSKLFFVFIAILC
Summary
Description
Catalyzes the dehydration of the S-form of NAD(P)HX at the expense of ATP, which is converted to ADP. Together with NAD(P)HX epimerase, which catalyzes the epimerization of the S-and R-forms, the enzyme allows the repair of both epimers of NAD(P)HX, a damaged form of NAD(P)H that is a result of enzymatic or heat-dependent hydration.
Required for the growth of imaginal tissues and for patterning of the adult wing.
Required for the growth of imaginal tissues and for patterning of the adult wing.
Catalytic Activity
(6S)-NADHX + ATP = ADP + H(+) + NADH + phosphate
Cofactor
Mg(2+)
Subunit
Homodimer; disulfide-linked.
Similarity
Belongs to the TGF-beta family.
Belongs to the NnrD/CARKD family.
Belongs to the NnrD/CARKD family.
Keywords
Cleavage on pair of basic residues
Complete proteome
Cytokine
Developmental protein
Disulfide bond
Glycoprotein
Growth factor
Reference proteome
Secreted
Signal
Feature
chain Protein 60A
Uniprot
H9J8U8
A0A0L7LT52
A0A1E1VXN5
A0A2H1WH31
A0A2W1BZ23
A0A2A4K4G5
+ More
A0A194RBI3 A0A194PRW2 A0A1Y1LUV6 A0A1Y1LQU8 D6WJG3 A0A1L8DJ25 A0A1B0GHD8 A0A195FVG3 F4X2S4 A0A151IUA6 A0A151WJZ7 A0A0L7RK70 A0A1B6EYZ3 A0A2A3EIB7 A0A088AQN8 A0A0C9R4S4 A0A1B6ISE8 A0A154NXB3 A0A158NLQ9 A0A026WF31 A0A1B6MMV4 A0A310S5S8 A0A195BLC4 A0A2Z6BDT4 A0A0J7KRT5 E9IW90 E2BSG9 N6UC73 N6TER8 E0VF83 W5U0I7 E2A4X2 A0A2P8ZE83 A0A336LTQ2 K7IQP9 A0A1J1HH35 A0A224XQP6 A0A2J7RPU7 A0A023F1G9 A0A034W7Y8 U5EVA1 A0A0K8WJZ0 A0A0A1WFZ2 A0A0A1XGQ1 A0A0A1X5T4 A0A084WIH8 W8C0F6 F8V237 A0A1I8NX77 A0A182RVG2 A0A0L0BNJ5 A0A067QZV7 Q7PZI7 A0A182HHM3 A0A182QT77 A0A182GRV7 T1IAN6 A0A1Q3FZB2 A0A182J0T4 A0A182WE14 A0A182XJR0 A0A182N2S5 A0A182L566 A0A182TGG5 A0A182V5A8 A0A182H371 A0A2R7W580 A0A023ERU8 A0A182SBB8 A0A182Y9I4 A0A1Q3FU05 A0A1Q3FU91 A0A182PCA3 T1PBW0 A0A1I8N5Z4 A0A182M031 Q292R9 A0A3Q0INX4 Q16WN3 A0A182FB81 A0A182KC53 A0A3B0IZ07 W5JLI6 B0XHY1 B0XHY0 B4PA79 A0A0B4LGA7 P27091 A0A1W4UZN3 B4I8U4 B4QB06
A0A194RBI3 A0A194PRW2 A0A1Y1LUV6 A0A1Y1LQU8 D6WJG3 A0A1L8DJ25 A0A1B0GHD8 A0A195FVG3 F4X2S4 A0A151IUA6 A0A151WJZ7 A0A0L7RK70 A0A1B6EYZ3 A0A2A3EIB7 A0A088AQN8 A0A0C9R4S4 A0A1B6ISE8 A0A154NXB3 A0A158NLQ9 A0A026WF31 A0A1B6MMV4 A0A310S5S8 A0A195BLC4 A0A2Z6BDT4 A0A0J7KRT5 E9IW90 E2BSG9 N6UC73 N6TER8 E0VF83 W5U0I7 E2A4X2 A0A2P8ZE83 A0A336LTQ2 K7IQP9 A0A1J1HH35 A0A224XQP6 A0A2J7RPU7 A0A023F1G9 A0A034W7Y8 U5EVA1 A0A0K8WJZ0 A0A0A1WFZ2 A0A0A1XGQ1 A0A0A1X5T4 A0A084WIH8 W8C0F6 F8V237 A0A1I8NX77 A0A182RVG2 A0A0L0BNJ5 A0A067QZV7 Q7PZI7 A0A182HHM3 A0A182QT77 A0A182GRV7 T1IAN6 A0A1Q3FZB2 A0A182J0T4 A0A182WE14 A0A182XJR0 A0A182N2S5 A0A182L566 A0A182TGG5 A0A182V5A8 A0A182H371 A0A2R7W580 A0A023ERU8 A0A182SBB8 A0A182Y9I4 A0A1Q3FU05 A0A1Q3FU91 A0A182PCA3 T1PBW0 A0A1I8N5Z4 A0A182M031 Q292R9 A0A3Q0INX4 Q16WN3 A0A182FB81 A0A182KC53 A0A3B0IZ07 W5JLI6 B0XHY1 B0XHY0 B4PA79 A0A0B4LGA7 P27091 A0A1W4UZN3 B4I8U4 B4QB06
EC Number
4.2.1.93
Pubmed
19121390
26227816
28756777
26354079
28004739
18362917
+ More
19820115 21719571 21347285 24508170 30249741 21282665 20798317 23537049 20566863 23595982 29403074 20075255 25474469 25348373 25830018 24438588 24495485 21627820 26108605 24845553 12364791 15302159 14747013 17210077 26483478 20966253 24945155 25244985 25315136 15632085 17994087 17510324 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1924384 1601181 9636086 22936249
19820115 21719571 21347285 24508170 30249741 21282665 20798317 23537049 20566863 23595982 29403074 20075255 25474469 25348373 25830018 24438588 24495485 21627820 26108605 24845553 12364791 15302159 14747013 17210077 26483478 20966253 24945155 25244985 25315136 15632085 17994087 17510324 20920257 23761445 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 1924384 1601181 9636086 22936249
EMBL
BABH01005042
JTDY01000181
KOB78411.1
GDQN01011569
JAT79485.1
ODYU01008632
+ More
SOQ52380.1 KZ149895 PZC78834.1 NWSH01000193 PCG78552.1 KQ460398 KPJ15193.1 KQ459601 KPI93875.1 GEZM01049224 JAV76030.1 GEZM01049225 JAV76029.1 KQ971342 EFA04645.1 GFDF01007616 JAV06468.1 AJWK01004477 AJWK01004478 KQ981219 KYN44416.1 GL888591 EGI59270.1 KQ980968 KYN10951.1 KQ983020 KYQ48199.1 KQ414578 KOC71208.1 GECZ01026602 JAS43167.1 KZ288253 PBC31022.1 GBYB01011279 JAG81046.1 GECU01017844 JAS89862.1 KQ434777 KZC04243.1 ADTU01019668 ADTU01019669 KK107238 QOIP01000007 EZA54725.1 RLU20214.1 GEBQ01002703 JAT37274.1 KQ769580 OAD52857.1 KQ976450 KYM85666.1 LC373544 BBD17787.1 LBMM01003898 KMQ93016.1 GL766458 EFZ15168.1 GL450233 EFN81357.1 APGK01040727 APGK01040728 KB740984 KB631904 ENN76252.1 ERL87081.1 ENN76253.1 DS235107 EEB12039.1 KC810052 AHH30785.1 GL436756 EFN71514.1 PYGN01000083 PSN54809.1 UFQS01000116 UFQT01000116 SSW99961.1 SSX20341.1 CVRI01000004 CRK87195.1 GFTR01005953 JAW10473.1 NEVH01001355 PNF42854.1 GBBI01003392 JAC15320.1 GAKP01008728 GAKP01008727 JAC50225.1 GANO01001967 JAB57904.1 GDHF01001134 JAI51180.1 GBXI01016666 JAC97625.1 GBXI01004155 JAD10137.1 GBXI01008061 JAD06231.1 ATLV01023934 KE525347 KFB50022.1 GAMC01007445 GAMC01007444 JAB99111.1 JN006973 AEI25992.1 JRES01001597 KNC21612.1 KK853149 KDR10681.1 AY578795 AAAB01008986 AAT07300.1 EAA00276.3 APCN01002428 AXCN02001087 JXUM01083317 KQ563369 KXJ73969.1 ACPB03006699 ACPB03006700 GFDL01002141 JAV32904.1 JXUM01106765 JXUM01106766 KQ565082 KXJ71365.1 KK854344 PTY14887.1 GAPW01001675 JAC11923.1 GFDL01004000 JAV31045.1 GFDL01003999 JAV31046.1 KA646169 AFP60798.1 AXCM01001233 CM000071 EAL24792.2 CH477560 EAT38991.1 OUUW01000001 SPP73147.1 ADMH02001211 ETN63639.1 DS233229 EDS28827.1 EDS28826.1 CM000158 EDW92405.1 AE013599 AHN56581.1 M77012 M84795 CH480824 EDW57019.1 CM000362 CM002911 EDX08443.1 KMY96132.1
SOQ52380.1 KZ149895 PZC78834.1 NWSH01000193 PCG78552.1 KQ460398 KPJ15193.1 KQ459601 KPI93875.1 GEZM01049224 JAV76030.1 GEZM01049225 JAV76029.1 KQ971342 EFA04645.1 GFDF01007616 JAV06468.1 AJWK01004477 AJWK01004478 KQ981219 KYN44416.1 GL888591 EGI59270.1 KQ980968 KYN10951.1 KQ983020 KYQ48199.1 KQ414578 KOC71208.1 GECZ01026602 JAS43167.1 KZ288253 PBC31022.1 GBYB01011279 JAG81046.1 GECU01017844 JAS89862.1 KQ434777 KZC04243.1 ADTU01019668 ADTU01019669 KK107238 QOIP01000007 EZA54725.1 RLU20214.1 GEBQ01002703 JAT37274.1 KQ769580 OAD52857.1 KQ976450 KYM85666.1 LC373544 BBD17787.1 LBMM01003898 KMQ93016.1 GL766458 EFZ15168.1 GL450233 EFN81357.1 APGK01040727 APGK01040728 KB740984 KB631904 ENN76252.1 ERL87081.1 ENN76253.1 DS235107 EEB12039.1 KC810052 AHH30785.1 GL436756 EFN71514.1 PYGN01000083 PSN54809.1 UFQS01000116 UFQT01000116 SSW99961.1 SSX20341.1 CVRI01000004 CRK87195.1 GFTR01005953 JAW10473.1 NEVH01001355 PNF42854.1 GBBI01003392 JAC15320.1 GAKP01008728 GAKP01008727 JAC50225.1 GANO01001967 JAB57904.1 GDHF01001134 JAI51180.1 GBXI01016666 JAC97625.1 GBXI01004155 JAD10137.1 GBXI01008061 JAD06231.1 ATLV01023934 KE525347 KFB50022.1 GAMC01007445 GAMC01007444 JAB99111.1 JN006973 AEI25992.1 JRES01001597 KNC21612.1 KK853149 KDR10681.1 AY578795 AAAB01008986 AAT07300.1 EAA00276.3 APCN01002428 AXCN02001087 JXUM01083317 KQ563369 KXJ73969.1 ACPB03006699 ACPB03006700 GFDL01002141 JAV32904.1 JXUM01106765 JXUM01106766 KQ565082 KXJ71365.1 KK854344 PTY14887.1 GAPW01001675 JAC11923.1 GFDL01004000 JAV31045.1 GFDL01003999 JAV31046.1 KA646169 AFP60798.1 AXCM01001233 CM000071 EAL24792.2 CH477560 EAT38991.1 OUUW01000001 SPP73147.1 ADMH02001211 ETN63639.1 DS233229 EDS28827.1 EDS28826.1 CM000158 EDW92405.1 AE013599 AHN56581.1 M77012 M84795 CH480824 EDW57019.1 CM000362 CM002911 EDX08443.1 KMY96132.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007266
+ More
UP000092461 UP000078541 UP000007755 UP000078492 UP000075809 UP000053825 UP000242457 UP000005203 UP000076502 UP000005205 UP000053097 UP000279307 UP000078540 UP000036403 UP000008237 UP000019118 UP000030742 UP000009046 UP000000311 UP000245037 UP000002358 UP000183832 UP000235965 UP000030765 UP000095300 UP000075900 UP000037069 UP000027135 UP000007062 UP000075840 UP000075886 UP000069940 UP000249989 UP000015103 UP000075880 UP000075920 UP000076407 UP000075884 UP000075882 UP000075902 UP000075903 UP000075901 UP000076408 UP000075885 UP000095301 UP000075883 UP000001819 UP000079169 UP000008820 UP000069272 UP000075881 UP000268350 UP000000673 UP000002320 UP000002282 UP000000803 UP000192221 UP000001292 UP000000304
UP000092461 UP000078541 UP000007755 UP000078492 UP000075809 UP000053825 UP000242457 UP000005203 UP000076502 UP000005205 UP000053097 UP000279307 UP000078540 UP000036403 UP000008237 UP000019118 UP000030742 UP000009046 UP000000311 UP000245037 UP000002358 UP000183832 UP000235965 UP000030765 UP000095300 UP000075900 UP000037069 UP000027135 UP000007062 UP000075840 UP000075886 UP000069940 UP000249989 UP000015103 UP000075880 UP000075920 UP000076407 UP000075884 UP000075882 UP000075902 UP000075903 UP000075901 UP000076408 UP000075885 UP000095301 UP000075883 UP000001819 UP000079169 UP000008820 UP000069272 UP000075881 UP000268350 UP000000673 UP000002320 UP000002282 UP000000803 UP000192221 UP000001292 UP000000304
Interpro
Gene 3D
ProteinModelPortal
H9J8U8
A0A0L7LT52
A0A1E1VXN5
A0A2H1WH31
A0A2W1BZ23
A0A2A4K4G5
+ More
A0A194RBI3 A0A194PRW2 A0A1Y1LUV6 A0A1Y1LQU8 D6WJG3 A0A1L8DJ25 A0A1B0GHD8 A0A195FVG3 F4X2S4 A0A151IUA6 A0A151WJZ7 A0A0L7RK70 A0A1B6EYZ3 A0A2A3EIB7 A0A088AQN8 A0A0C9R4S4 A0A1B6ISE8 A0A154NXB3 A0A158NLQ9 A0A026WF31 A0A1B6MMV4 A0A310S5S8 A0A195BLC4 A0A2Z6BDT4 A0A0J7KRT5 E9IW90 E2BSG9 N6UC73 N6TER8 E0VF83 W5U0I7 E2A4X2 A0A2P8ZE83 A0A336LTQ2 K7IQP9 A0A1J1HH35 A0A224XQP6 A0A2J7RPU7 A0A023F1G9 A0A034W7Y8 U5EVA1 A0A0K8WJZ0 A0A0A1WFZ2 A0A0A1XGQ1 A0A0A1X5T4 A0A084WIH8 W8C0F6 F8V237 A0A1I8NX77 A0A182RVG2 A0A0L0BNJ5 A0A067QZV7 Q7PZI7 A0A182HHM3 A0A182QT77 A0A182GRV7 T1IAN6 A0A1Q3FZB2 A0A182J0T4 A0A182WE14 A0A182XJR0 A0A182N2S5 A0A182L566 A0A182TGG5 A0A182V5A8 A0A182H371 A0A2R7W580 A0A023ERU8 A0A182SBB8 A0A182Y9I4 A0A1Q3FU05 A0A1Q3FU91 A0A182PCA3 T1PBW0 A0A1I8N5Z4 A0A182M031 Q292R9 A0A3Q0INX4 Q16WN3 A0A182FB81 A0A182KC53 A0A3B0IZ07 W5JLI6 B0XHY1 B0XHY0 B4PA79 A0A0B4LGA7 P27091 A0A1W4UZN3 B4I8U4 B4QB06
A0A194RBI3 A0A194PRW2 A0A1Y1LUV6 A0A1Y1LQU8 D6WJG3 A0A1L8DJ25 A0A1B0GHD8 A0A195FVG3 F4X2S4 A0A151IUA6 A0A151WJZ7 A0A0L7RK70 A0A1B6EYZ3 A0A2A3EIB7 A0A088AQN8 A0A0C9R4S4 A0A1B6ISE8 A0A154NXB3 A0A158NLQ9 A0A026WF31 A0A1B6MMV4 A0A310S5S8 A0A195BLC4 A0A2Z6BDT4 A0A0J7KRT5 E9IW90 E2BSG9 N6UC73 N6TER8 E0VF83 W5U0I7 E2A4X2 A0A2P8ZE83 A0A336LTQ2 K7IQP9 A0A1J1HH35 A0A224XQP6 A0A2J7RPU7 A0A023F1G9 A0A034W7Y8 U5EVA1 A0A0K8WJZ0 A0A0A1WFZ2 A0A0A1XGQ1 A0A0A1X5T4 A0A084WIH8 W8C0F6 F8V237 A0A1I8NX77 A0A182RVG2 A0A0L0BNJ5 A0A067QZV7 Q7PZI7 A0A182HHM3 A0A182QT77 A0A182GRV7 T1IAN6 A0A1Q3FZB2 A0A182J0T4 A0A182WE14 A0A182XJR0 A0A182N2S5 A0A182L566 A0A182TGG5 A0A182V5A8 A0A182H371 A0A2R7W580 A0A023ERU8 A0A182SBB8 A0A182Y9I4 A0A1Q3FU05 A0A1Q3FU91 A0A182PCA3 T1PBW0 A0A1I8N5Z4 A0A182M031 Q292R9 A0A3Q0INX4 Q16WN3 A0A182FB81 A0A182KC53 A0A3B0IZ07 W5JLI6 B0XHY1 B0XHY0 B4PA79 A0A0B4LGA7 P27091 A0A1W4UZN3 B4I8U4 B4QB06
Ontologies
GO
GO:0016021
GO:0008083
GO:0005576
GO:0005125
GO:0010862
GO:0070700
GO:0048468
GO:0005160
GO:0043408
GO:0060395
GO:0030509
GO:0042981
GO:0005615
GO:0052855
GO:0047453
GO:0005524
GO:0046496
GO:0046928
GO:0008586
GO:0007474
GO:0031045
GO:0097467
GO:0045887
GO:0036099
GO:0007274
GO:0007528
GO:0099504
GO:0035222
GO:0048636
GO:0030707
GO:0012505
GO:0007419
GO:0007391
GO:0040007
GO:0015074
GO:0008026
GO:0003723
GO:0043401
GO:0003677
GO:0003707
GO:0004651
GO:0005634
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 21,
Likelihood: 0.794294
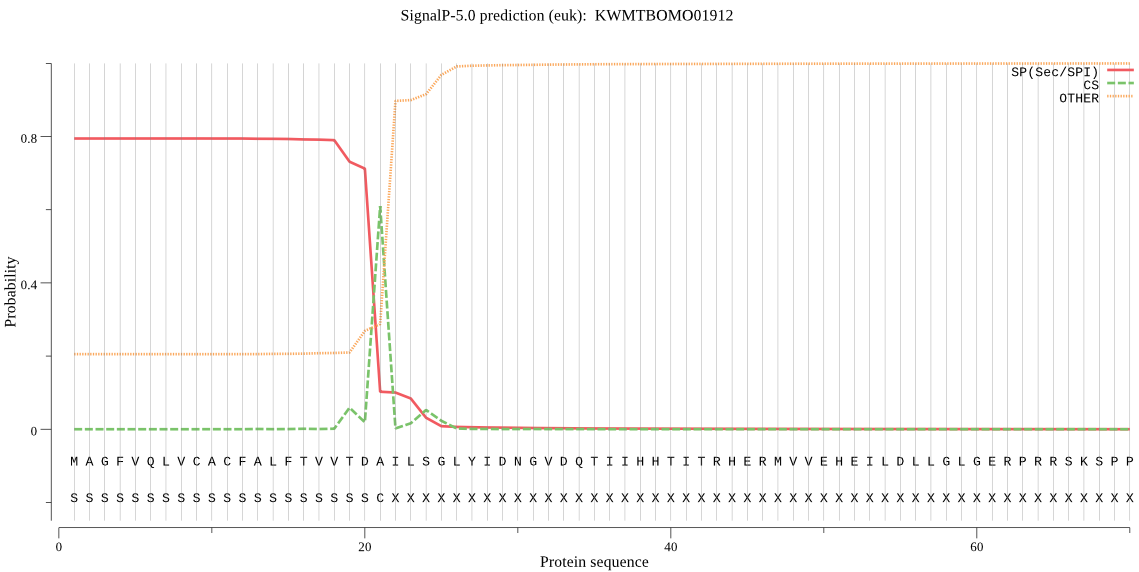
Length:
135
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.47895
Exp number, first 60 AAs:
20.41454
Total prob of N-in:
0.32313
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 26
inside
27 - 135

Population Genetic Test Statistics
Pi
10.075078
Theta
14.00891
Tajima's D
-0.937512
CLR
574.647327
CSRT
0.146742662866857
Interpretation
Uncertain
