Gene
KWMTBOMO01909
Pre Gene Modal
BGIBMGA005941
Annotation
PREDICTED:_G-protein_coupled_receptor_Mth_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.016
Sequence
CDS
ATGACGTGGCATTTGTTCTTTATTTGCGTATTTTTGTATTATACAAGTGTCAGTGCTGAATCCTATGAAAAAGTGGTTGCATTTAGGAAGTGTTGTATTGAGAATCAAAGTTTAGTTAGATCATTTGAAGTCAACAAGACTGAAAGTTTTGTATGCCTAGACCGGTCCACTATTGAAGATAATTATAATATTTCTGCATCTCCACTCCTGATAGGTGAGAATGTTAATACTGTATATGGCACCCCGGTGTGTGACCAGATGGAATTGTCTCAAGTGACTGCTACTGAATTCGATTCTGATCAAGACCATCGTTGCTATGACAGACTGATAATAGAAACGATAAACGGCACGATTAAACAGAACATACCGAAAATTGTTGCTTTAACTTGCGCGAATTTTAAAAATAATACGAAGAATCTCATGAAATCAAAGCTAACGATAGATCGCATCAAGAAATGTTGTCCAAAAGGTCAAAGCTACGATCCAAAATATCATTTTTGTAGGCTCAGCGACTCGGAAAATAATGAACAGGCGCTAATAAAAAGATTAAATTTGAGTTCAGATCATATTTATGAAGTTGAGAACGAGTTACATTGTAAAGCTGTTGAAACCGGTATTGAATTGAGTGAAGAGAAATTTTCGTTAGTAGTAGAAGGAAGTAACTTAATTGTGACCAGAAAATTTTCAGAAAAGAGTCATATTGCACACCCCGGAGAATGGTGCATAGATCGCATGTTCCCCGGGGAAAGTCTCATTGCTCGAATGTGCACAAACAGCTGTTCTGATTTTGATGCGTTTTGTGTGAAGAAGTGTTGTCCTGTGGGACAGCATTATAAAACGTTGCGATGCGGAAGTTTTGTCAGTAGATGTGTGCCGAATAAGGATAGCGTGGCGTTTAATATTTCGAATTATTTGGACGAATTAAAGGCTAATTATGATGATATCCCGGATGTGCTTGGGATCCGTTCAGACTTGCTGTGCTCCGCTGGAAGAGTGGCACTAAACATAAGTGCTGATTATGACAAACACGAATTGACTCGAGATGGATATCTTCGGTCTGCTATAAGCGATCATGGTGATTACTGTTTTGAGACATTCGATAGTAGACAGTGTCCTGGTGGTGATTTGACCGTTACCGCAGTCACATGTTTCGTCAAACCGCAGATGAAAAATTTCCAAGTATCTTTTTTGCTTATTAGCATATCGAGCGTATGTCTGGCATTGACGTTGTTGGTGTATTGCGCTTTGCCAGAATTGTGGAACCTACACGGTCGCACCTTAATCTGCCACGTTAGTATGATGTTGCTGGCTTTCACCTGCCTCGCAAGGGTTCAGTATAAAGAAGTTGCTGATAATAAACTCTGTACATTGCTGGGGTACGGGATCTACTTTGGATTCGTAGCGGCATTTGCTTGGCTTAACGTCATGTGCCTCGACATCTGGTGGACATTTGGGAGTGTGAGAATGGTGCAACCTTTGAGGAAGTCAGGAGCCGAGTGGCGAAGATTTCTCTGGTATTCGTTGAATGCGTGGGGTTTTGCGAGCATCCTGACCCTTGTAATGTTTATTCTCGACGAGTATCCTATTGCAATAGAACTTGACGCCAACATCGGCAGCGGCATGTGTTGGTTTGGCTCACTACAAAACGAGCGGTCAGATTGGCCTCACTACATTTTCTTCGTGATTCCGATGGGTCTTGTGACATGTACGAATTTCATCCTGTGGGTGTTGACGGCGCGGCATTGCGCTCGAGTCAAGTCCGAGGTCCATCGGCTGCAGGCGGGCTCGGTTGGTGACCGCGCCAAGCGACGTTTCCGTATTGACAGAGCTAAGTACCTTTTGACCGGAAAACTGTGGGTGGTGATGGGAGCGGGATGGATATCGGAGCTTCTTTCTACTATCGCTTCAGAACCAAAGTGGCTTTGGACCATCGTTGACCTTCTGAATGAACTACAAGGAGTCTTCATATTCCTCATACTAGTGTTCAAGCCTAAGCTGTACTACCTAATCCGAAAGAGACTAGGGCTGGAGAAACCGGATGCTCAGAAGAATGGCGGTACGTCATCTGGTCGTACATCTTCCACCTTTCTGTCGAGGACCATCAGCAATGACGAGCGCGCCAATCTCCGAGTTTCACAACTTAACACCAAGCAAGCTTAG
Protein
MTWHLFFICVFLYYTSVSAESYEKVVAFRKCCIENQSLVRSFEVNKTESFVCLDRSTIEDNYNISASPLLIGENVNTVYGTPVCDQMELSQVTATEFDSDQDHRCYDRLIIETINGTIKQNIPKIVALTCANFKNNTKNLMKSKLTIDRIKKCCPKGQSYDPKYHFCRLSDSENNEQALIKRLNLSSDHIYEVENELHCKAVETGIELSEEKFSLVVEGSNLIVTRKFSEKSHIAHPGEWCIDRMFPGESLIARMCTNSCSDFDAFCVKKCCPVGQHYKTLRCGSFVSRCVPNKDSVAFNISNYLDELKANYDDIPDVLGIRSDLLCSAGRVALNISADYDKHELTRDGYLRSAISDHGDYCFETFDSRQCPGGDLTVTAVTCFVKPQMKNFQVSFLLISISSVCLALTLLVYCALPELWNLHGRTLICHVSMMLLAFTCLARVQYKEVADNKLCTLLGYGIYFGFVAAFAWLNVMCLDIWWTFGSVRMVQPLRKSGAEWRRFLWYSLNAWGFASILTLVMFILDEYPIAIELDANIGSGMCWFGSLQNERSDWPHYIFFVIPMGLVTCTNFILWVLTARHCARVKSEVHRLQAGSVGDRAKRRFRIDRAKYLLTGKLWVVMGAGWISELLSTIASEPKWLWTIVDLLNELQGVFIFLILVFKPKLYYLIRKRLGLEKPDAQKNGGTSSGRTSSTFLSRTISNDERANLRVSQLNTKQA
Summary
Uniprot
A0A0U1ZWU3
A0A2W1BYV7
A0A194PKM2
A0A0L7LC34
A0A212ETW6
A0A1E1WU03
+ More
A0A2H1WH34 A0A2P8ZCL8 A0A1L8DJQ8 A0A195EU48 A0A195E5N6 A0A195BA00 A0A2J7RJJ1 A0A158NGK2 F4WP60 A0A151IKT3 A0A151WM86 A0A1I8PMX5 E2A3J4 A0A026WP22 A0A3L8DYN1 A0A154NYP2 A0A1J1IM80 E2BKF8 A0A1I8N3N5 A0A0M4E569 A0A0J9QZV4 B3N944 A0A1B6MLQ0 B4Q8R3 J9K1T3 B4MG46 A0A0Q9W024 B4NZ68 A0A0R1DL55 A0A034W2A7 A0A1W4USW0 B4HWI0 A0A1Y1LC53 E2QCS3 A0A0M8ZUT3 A0A2H8TQ57 W8BB25 A0A336MI57 A0A2S2Q9I7 A0A336MEK8 A0A232FE72 A0A0A9XPA3 A0A336KQJ1 A0A0P9BPZ4 A0A336KTJ3 A0A1A9WBV7 A0A1A9UDD9 A0A2Z5H022 E0VFN2 J9KXA2 A0A0P4XXW6 A0A0P5JBW7 J9K255
A0A2H1WH34 A0A2P8ZCL8 A0A1L8DJQ8 A0A195EU48 A0A195E5N6 A0A195BA00 A0A2J7RJJ1 A0A158NGK2 F4WP60 A0A151IKT3 A0A151WM86 A0A1I8PMX5 E2A3J4 A0A026WP22 A0A3L8DYN1 A0A154NYP2 A0A1J1IM80 E2BKF8 A0A1I8N3N5 A0A0M4E569 A0A0J9QZV4 B3N944 A0A1B6MLQ0 B4Q8R3 J9K1T3 B4MG46 A0A0Q9W024 B4NZ68 A0A0R1DL55 A0A034W2A7 A0A1W4USW0 B4HWI0 A0A1Y1LC53 E2QCS3 A0A0M8ZUT3 A0A2H8TQ57 W8BB25 A0A336MI57 A0A2S2Q9I7 A0A336MEK8 A0A232FE72 A0A0A9XPA3 A0A336KQJ1 A0A0P9BPZ4 A0A336KTJ3 A0A1A9WBV7 A0A1A9UDD9 A0A2Z5H022 E0VFN2 J9KXA2 A0A0P4XXW6 A0A0P5JBW7 J9K255
Pubmed
EMBL
KM205446
AKA95280.1
KZ149895
PZC78835.1
KQ459601
KPI93877.1
+ More
JTDY01001766 KOB72950.1 AGBW02012530 OWR44933.1 GDQN01000525 JAT90529.1 ODYU01008632 SOQ52379.1 PYGN01000104 PSN54243.1 GFDF01007386 JAV06698.1 KQ981965 KYN31773.1 KQ979608 KYN20401.1 KQ976542 KYM81024.1 NEVH01002992 PNF40994.1 ADTU01015065 GL888243 EGI63991.1 KQ977164 KYN05227.1 KQ982944 KYQ48940.1 GL436444 EFN71976.1 KK107139 EZA57807.1 QOIP01000003 RLU25225.1 KQ434777 KZC04194.1 CVRI01000055 CRL01349.1 GL448794 EFN83825.1 CP012523 ALC39196.1 CM002910 KMY89441.1 CH954177 EDV58479.2 GEBQ01003133 JAT36844.1 CM000361 EDX04478.1 ABLF02022044 ABLF02022046 ABLF02028129 CH940671 EDW58184.2 KRF78166.1 CM000157 EDW88763.2 KRJ98015.1 GAKP01010470 JAC48482.1 CH480818 EDW52375.1 GEZM01060140 JAV71249.1 AE014134 AAN10727.3 KQ435861 KOX70436.1 GFXV01004522 MBW16327.1 GAMC01010658 JAB95897.1 UFQT01000920 SSX28017.1 GGMS01005017 MBY74220.1 UFQS01000718 UFQT01000718 SSX06404.1 SSX26757.1 NNAY01000372 OXU28823.1 GBHO01021800 JAG21804.1 UFQS01000704 UFQT01000704 SSX06311.1 SSX26665.1 CH902620 KPU73881.1 SSX06403.1 SSX26756.1 MH175212 AXC32920.1 DS235123 EEB12188.1 ABLF02015080 GDIP01245006 JAI78395.1 GDIQ01227244 JAK24481.1 ABLF02023318
JTDY01001766 KOB72950.1 AGBW02012530 OWR44933.1 GDQN01000525 JAT90529.1 ODYU01008632 SOQ52379.1 PYGN01000104 PSN54243.1 GFDF01007386 JAV06698.1 KQ981965 KYN31773.1 KQ979608 KYN20401.1 KQ976542 KYM81024.1 NEVH01002992 PNF40994.1 ADTU01015065 GL888243 EGI63991.1 KQ977164 KYN05227.1 KQ982944 KYQ48940.1 GL436444 EFN71976.1 KK107139 EZA57807.1 QOIP01000003 RLU25225.1 KQ434777 KZC04194.1 CVRI01000055 CRL01349.1 GL448794 EFN83825.1 CP012523 ALC39196.1 CM002910 KMY89441.1 CH954177 EDV58479.2 GEBQ01003133 JAT36844.1 CM000361 EDX04478.1 ABLF02022044 ABLF02022046 ABLF02028129 CH940671 EDW58184.2 KRF78166.1 CM000157 EDW88763.2 KRJ98015.1 GAKP01010470 JAC48482.1 CH480818 EDW52375.1 GEZM01060140 JAV71249.1 AE014134 AAN10727.3 KQ435861 KOX70436.1 GFXV01004522 MBW16327.1 GAMC01010658 JAB95897.1 UFQT01000920 SSX28017.1 GGMS01005017 MBY74220.1 UFQS01000718 UFQT01000718 SSX06404.1 SSX26757.1 NNAY01000372 OXU28823.1 GBHO01021800 JAG21804.1 UFQS01000704 UFQT01000704 SSX06311.1 SSX26665.1 CH902620 KPU73881.1 SSX06403.1 SSX26756.1 MH175212 AXC32920.1 DS235123 EEB12188.1 ABLF02015080 GDIP01245006 JAI78395.1 GDIQ01227244 JAK24481.1 ABLF02023318
Proteomes
UP000053268
UP000037510
UP000007151
UP000245037
UP000078541
UP000078492
+ More
UP000078540 UP000235965 UP000005205 UP000007755 UP000078542 UP000075809 UP000095300 UP000000311 UP000053097 UP000279307 UP000076502 UP000183832 UP000008237 UP000095301 UP000092553 UP000008711 UP000000304 UP000007819 UP000008792 UP000002282 UP000192221 UP000001292 UP000000803 UP000053105 UP000215335 UP000007801 UP000091820 UP000078200 UP000009046
UP000078540 UP000235965 UP000005205 UP000007755 UP000078542 UP000075809 UP000095300 UP000000311 UP000053097 UP000279307 UP000076502 UP000183832 UP000008237 UP000095301 UP000092553 UP000008711 UP000000304 UP000007819 UP000008792 UP000002282 UP000192221 UP000001292 UP000000803 UP000053105 UP000215335 UP000007801 UP000091820 UP000078200 UP000009046
Interpro
SUPFAM
SSF63877
SSF63877
Gene 3D
ProteinModelPortal
A0A0U1ZWU3
A0A2W1BYV7
A0A194PKM2
A0A0L7LC34
A0A212ETW6
A0A1E1WU03
+ More
A0A2H1WH34 A0A2P8ZCL8 A0A1L8DJQ8 A0A195EU48 A0A195E5N6 A0A195BA00 A0A2J7RJJ1 A0A158NGK2 F4WP60 A0A151IKT3 A0A151WM86 A0A1I8PMX5 E2A3J4 A0A026WP22 A0A3L8DYN1 A0A154NYP2 A0A1J1IM80 E2BKF8 A0A1I8N3N5 A0A0M4E569 A0A0J9QZV4 B3N944 A0A1B6MLQ0 B4Q8R3 J9K1T3 B4MG46 A0A0Q9W024 B4NZ68 A0A0R1DL55 A0A034W2A7 A0A1W4USW0 B4HWI0 A0A1Y1LC53 E2QCS3 A0A0M8ZUT3 A0A2H8TQ57 W8BB25 A0A336MI57 A0A2S2Q9I7 A0A336MEK8 A0A232FE72 A0A0A9XPA3 A0A336KQJ1 A0A0P9BPZ4 A0A336KTJ3 A0A1A9WBV7 A0A1A9UDD9 A0A2Z5H022 E0VFN2 J9KXA2 A0A0P4XXW6 A0A0P5JBW7 J9K255
A0A2H1WH34 A0A2P8ZCL8 A0A1L8DJQ8 A0A195EU48 A0A195E5N6 A0A195BA00 A0A2J7RJJ1 A0A158NGK2 F4WP60 A0A151IKT3 A0A151WM86 A0A1I8PMX5 E2A3J4 A0A026WP22 A0A3L8DYN1 A0A154NYP2 A0A1J1IM80 E2BKF8 A0A1I8N3N5 A0A0M4E569 A0A0J9QZV4 B3N944 A0A1B6MLQ0 B4Q8R3 J9K1T3 B4MG46 A0A0Q9W024 B4NZ68 A0A0R1DL55 A0A034W2A7 A0A1W4USW0 B4HWI0 A0A1Y1LC53 E2QCS3 A0A0M8ZUT3 A0A2H8TQ57 W8BB25 A0A336MI57 A0A2S2Q9I7 A0A336MEK8 A0A232FE72 A0A0A9XPA3 A0A336KQJ1 A0A0P9BPZ4 A0A336KTJ3 A0A1A9WBV7 A0A1A9UDD9 A0A2Z5H022 E0VFN2 J9KXA2 A0A0P4XXW6 A0A0P5JBW7 J9K255
Ontologies
GO
Topology
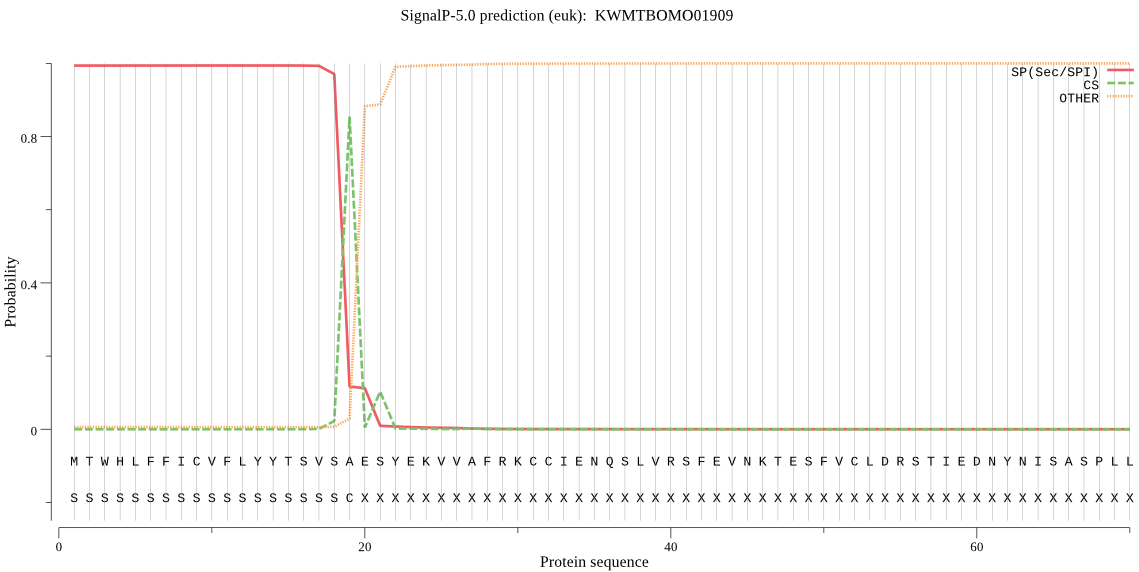
SignalP
Position: 1 - 19,
Likelihood: 0.993536
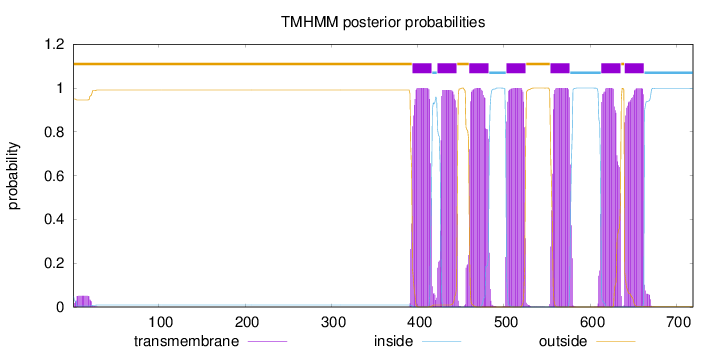
Length:
719
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
154.87853
Exp number, first 60 AAs:
0.93503
Total prob of N-in:
0.05007
outside
1 - 393
TMhelix
394 - 416
inside
417 - 422
TMhelix
423 - 445
outside
446 - 459
TMhelix
460 - 482
inside
483 - 502
TMhelix
503 - 525
outside
526 - 553
TMhelix
554 - 576
inside
577 - 612
TMhelix
613 - 635
outside
636 - 639
TMhelix
640 - 662
inside
663 - 719
Population Genetic Test Statistics
Pi
244.123302
Theta
195.009564
Tajima's D
0.842097
CLR
0.061073
CSRT
0.611069446527674
Interpretation
Uncertain
