Gene
KWMTBOMO01905
Pre Gene Modal
BGIBMGA006204
Annotation
PREDICTED:_mitochondrial_sodium/hydrogen_exchanger_9B2-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.142
Sequence
CDS
ATGAACGAAGAAGACTGCTTTATAGTCAGGATAGCAGACGACGTTAAGGACAGTATGAATCAATCACTGCCAGACTTGTCACCTAGTACCGATACCGAAAAAAATACCTCAAAGGATAATGTGAAAAATAAAGGTACTTGGTTACATACATCGACTCCAACAGTTGTTCATTTTTTGGCATTAATTACTCAAGGTCTTTTAGTTTGGGGGCTACTTTGGCTTGCGTGGGGAGACGGGTGGAGTTGGAATGGAAAATGGTTCAAACCAGCAGTCGTTGTGGTTATCGCATGGAGCAGCGGACAACTCCTACAGAGACTCACTACGCTCCCTCCACTTCTGGCTGCTATACTAACAGGAATCCTTGCAAGAAATTTAGGCTACCTCGACATGCGAGGATACCCACACGTGGACAACTTTTTAAGGAAAATTTACCCAGTGATAATTCTTGGCAAGTCATCTCTGGGTTGGGACTTGAAATTTATGAAGGCCAATTGGCGTCAAGTTTTCAGCCTCGGTGTGGTACCCTGGACGGCTGAGGTTCTCGCTCTCGCGACATGCTCGCGGTACTTCCTCCATTATCCCTGGGTTTGGGGCATTCTCCTTGGGTCTATCTACGCATCTTTGTCTTGTGCTGTGGTGATGCCCTCTGTATTAAAAGTTGGGACTAGTCCAGATAGGACCCACAACTGGCCTCAGTTGTGCTCGACTGCCGGAGGAATCGATACTGCACTCAGCGTTGGAATATATGGGATCGTATATAGCTACATTTTCAGTGATGTGCACGATGCTTATCGATACACTAAAGTGGCGTTGACCTTATTCGTCGGAGTCGCTATGGGTATCGCTTGGGGCAGTCTCGCAAAATTGATACCTCACTCTCGAAACCCCTACGTTACTGAATTGAGGGTTCTCTTCGTGTTACTCGGTGGACTGTTTGGAAACTTTTTGACGCTGCACTTGGGCTGGGGCGGCGCTGGCGGTGTAGCTGTATTAGCGTGCAACGCAACAGCGGCGAATCACTGGTCGCGCGACGGTTGGAAGTTAAACAACAACCCTGCCTCGACCGTTTATCGCGTTTTGTGGGCAGCTTTGGAGCCTGCCGTGTTCGCATACACTGGAACATACTTTGAAATACAACACACATTAACTGACACAATACTAAAAGGATTTGGAATCCTGTTAGTTTGTTTAGCTGTGAGACTAGCGTGCGCTTTCCTGATGTGTTGGGATATGAGCGTCAAGGAAAGAATTTTTATATGCTGCAGCTGGACGCCAAAATCTATAGTGGAGGCCGTACTATGTCCGCTGGCACTGAACACATTAATTTTACGAGGCGGAAAAAACGAACAAGAAATGGAATACGCAGAGCAACTAATGAGGCTAGTTGTTCAAGCAATTCTAGTAACAACTCCTGTTGGTTATTTGTTCACAAAGCATTTGGGGCCGGTGCTGTTACGAGACAAACGCAAAGAATTTCGGAACGAGTCCGAAGCATGA
Protein
MNEEDCFIVRIADDVKDSMNQSLPDLSPSTDTEKNTSKDNVKNKGTWLHTSTPTVVHFLALITQGLLVWGLLWLAWGDGWSWNGKWFKPAVVVVIAWSSGQLLQRLTTLPPLLAAILTGILARNLGYLDMRGYPHVDNFLRKIYPVIILGKSSLGWDLKFMKANWRQVFSLGVVPWTAEVLALATCSRYFLHYPWVWGILLGSIYASLSCAVVMPSVLKVGTSPDRTHNWPQLCSTAGGIDTALSVGIYGIVYSYIFSDVHDAYRYTKVALTLFVGVAMGIAWGSLAKLIPHSRNPYVTELRVLFVLLGGLFGNFLTLHLGWGGAGGVAVLACNATAANHWSRDGWKLNNNPASTVYRVLWAALEPAVFAYTGTYFEIQHTLTDTILKGFGILLVCLAVRLACAFLMCWDMSVKERIFICCSWTPKSIVEAVLCPLALNTLILRGGKNEQEMEYAEQLMRLVVQAILVTTPVGYLFTKHLGPVLLRDKRKEFRNESEA
Summary
Uniprot
H9J9L1
A0A2A4K313
A0A2H1WQE8
A0A2W1BUW0
A0A194RD48
A0A194PKB6
+ More
A0A212FG08 A0A0L7LSH2 A0A212FG10 A0A2W1BV43 A0A2A4K4S2 A0A194RGZ3 T1DU38 A0A182FRV9 A0A182T952 W5JHL6 A0A182VRG1 A0A182NFP2 A0A182RX38 A0A182M6L2 A0MNP7 A0A182XGF3 A0A182HZX3 A0A182U2G4 Q7PYT2 A0A182QVG0 A0A182JL69 A0A182UNV4 A0A182XW09 A0A182LR05 A0A084WSH3 A0A182PE22 A0A2J7RJI7 A0A026WQ33 T1PB54 A0A1I8MUY9 A0A1I8MUZ4 A0A2A4JX14 B4KJX2 B0WSL1 A0A1B6K4L7 A0A194PJ27 A0A194R0K4 A0A0M4E945 A0A1Q3FZQ1 A0A1A9WXC6 A0A1B6IHG3 A0A0Q9X3C6 A0A0J7L4S6 A0A3B0JGT3 A0A0Q9X184 B4N076 B4LRP8 Q95U27 Q16R10 H9J9L4 A0A0R3NVL1 A0A2H1VLY0 F2FB79 A0A1L8DDK3 B4GKY8 Q29JY6 Q9VM74 A0A0R3NV36 A0A023EVB6 Q7KTL6 A0A3B0K7E6 A0A3B0JHA4 A0A0J9QX13 A0A0J9QXC9 A0A1L8DE11 A0A0Q5WNB0 A0A0Q5WK11 B4LRP9 B3N5Y2 A0A0Q9WAQ8 B4JQL1 A0A0A1XLT2 B4HXY9 A0A0A1XIM4 A0A212FGB7 A0A1L8E003 A0A1L8E028 A0A0R1DKJ9 B4P000 A0A1W4UNP4 A0A1L8DZT1 A0A1W4UNX2 A0A1I8NMR4 A0A1I8NMR0 E0VFM9 E2A3J5 A0A0P9BNS5 A0A1Y1NIF1 B3MKQ9 A0A232FET6 A0A1Y1NMV7 A0A1Y1NLM9 A0A1Y1NJI0
A0A212FG08 A0A0L7LSH2 A0A212FG10 A0A2W1BV43 A0A2A4K4S2 A0A194RGZ3 T1DU38 A0A182FRV9 A0A182T952 W5JHL6 A0A182VRG1 A0A182NFP2 A0A182RX38 A0A182M6L2 A0MNP7 A0A182XGF3 A0A182HZX3 A0A182U2G4 Q7PYT2 A0A182QVG0 A0A182JL69 A0A182UNV4 A0A182XW09 A0A182LR05 A0A084WSH3 A0A182PE22 A0A2J7RJI7 A0A026WQ33 T1PB54 A0A1I8MUY9 A0A1I8MUZ4 A0A2A4JX14 B4KJX2 B0WSL1 A0A1B6K4L7 A0A194PJ27 A0A194R0K4 A0A0M4E945 A0A1Q3FZQ1 A0A1A9WXC6 A0A1B6IHG3 A0A0Q9X3C6 A0A0J7L4S6 A0A3B0JGT3 A0A0Q9X184 B4N076 B4LRP8 Q95U27 Q16R10 H9J9L4 A0A0R3NVL1 A0A2H1VLY0 F2FB79 A0A1L8DDK3 B4GKY8 Q29JY6 Q9VM74 A0A0R3NV36 A0A023EVB6 Q7KTL6 A0A3B0K7E6 A0A3B0JHA4 A0A0J9QX13 A0A0J9QXC9 A0A1L8DE11 A0A0Q5WNB0 A0A0Q5WK11 B4LRP9 B3N5Y2 A0A0Q9WAQ8 B4JQL1 A0A0A1XLT2 B4HXY9 A0A0A1XIM4 A0A212FGB7 A0A1L8E003 A0A1L8E028 A0A0R1DKJ9 B4P000 A0A1W4UNP4 A0A1L8DZT1 A0A1W4UNX2 A0A1I8NMR4 A0A1I8NMR0 E0VFM9 E2A3J5 A0A0P9BNS5 A0A1Y1NIF1 B3MKQ9 A0A232FET6 A0A1Y1NMV7 A0A1Y1NLM9 A0A1Y1NJI0
Pubmed
19121390
28756777
26354079
22118469
26227816
20920257
+ More
23761445 17951426 12364791 14747013 17210077 25244985 20966253 24438588 24508170 30249741 25315136 17994087 17510324 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24945155 26109357 26109356 22936249 25830018 17550304 20566863 20798317 28004739 28648823
23761445 17951426 12364791 14747013 17210077 25244985 20966253 24438588 24508170 30249741 25315136 17994087 17510324 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24945155 26109357 26109356 22936249 25830018 17550304 20566863 20798317 28004739 28648823
EMBL
BABH01005033
NWSH01000184
PCG78651.1
ODYU01010224
SOQ55196.1
KZ149895
+ More
PZC78838.1 KQ460398 KPJ15200.1 KQ459601 KPI93881.1 AGBW02008736 OWR52668.1 JTDY01000180 KOB78423.1 OWR52669.1 PZC78839.1 PCG78650.1 KPJ15201.1 GAMD01000494 JAB01097.1 ADMH02001227 ETN63586.1 AXCM01001046 EF014219 ABJ91581.1 APCN01002017 APCN01002018 AAAB01008987 EAA01006.3 AXCN02000239 ATLV01026539 ATLV01026540 ATLV01026541 ATLV01026542 ATLV01026543 KE525415 KFB53167.1 NEVH01002992 PNF40993.1 KK107139 QOIP01000003 EZA57806.1 RLU24744.1 KA645380 AFP60009.1 NWSH01000511 PCG75950.1 CH933807 EDW12575.1 DS232073 EDS33958.1 GECU01001319 JAT06388.1 KQ459603 KPI92724.1 KQ460883 KPJ11232.1 CP012523 ALC38651.1 GFDL01002000 JAV33045.1 GECU01035477 GECU01021350 JAS72229.1 JAS86356.1 CH963920 KRF98791.1 LBMM01000631 KMQ97942.1 OUUW01000006 SPP81567.1 KRF98790.1 EDW78011.2 CH940649 EDW64650.1 AY058354 AAL13583.1 CH477726 EAT36864.1 BABH01005037 CH379062 KRT04988.1 ODYU01003105 SOQ41442.1 BT126166 ADZ99418.1 GFDF01009531 JAV04553.1 CH479184 EDW37304.1 EAL32825.3 AE014134 AAF52449.2 KRT04987.1 GAPW01000535 JAC13063.1 BT031332 AAN10603.1 ABY21745.1 SPP81566.1 SPP81565.1 CM002910 KMY88637.1 KMY88638.1 GFDF01009530 JAV04554.1 CH954177 KQS70650.1 KQS70651.1 EDW64651.1 EDV59141.2 KRF81772.1 CH916372 EDV99191.1 GBXI01002321 JAD11971.1 CH480818 EDW51919.1 GBXI01003929 JAD10363.1 AGBW02008689 OWR52770.1 GFDF01002073 JAV12011.1 GFDF01002075 JAV12009.1 CM000157 KRJ97422.1 EDW87827.2 GFDF01002074 JAV12010.1 DS235123 EEB12185.1 GL436444 EFN71977.1 CH902620 KPU73497.1 GEZM01001569 JAV97664.1 EDV31590.2 NNAY01000372 OXU28817.1 GEZM01001567 JAV97666.1 GEZM01001568 GEZM01001566 JAV97665.1 GEZM01001565 JAV97668.1
PZC78838.1 KQ460398 KPJ15200.1 KQ459601 KPI93881.1 AGBW02008736 OWR52668.1 JTDY01000180 KOB78423.1 OWR52669.1 PZC78839.1 PCG78650.1 KPJ15201.1 GAMD01000494 JAB01097.1 ADMH02001227 ETN63586.1 AXCM01001046 EF014219 ABJ91581.1 APCN01002017 APCN01002018 AAAB01008987 EAA01006.3 AXCN02000239 ATLV01026539 ATLV01026540 ATLV01026541 ATLV01026542 ATLV01026543 KE525415 KFB53167.1 NEVH01002992 PNF40993.1 KK107139 QOIP01000003 EZA57806.1 RLU24744.1 KA645380 AFP60009.1 NWSH01000511 PCG75950.1 CH933807 EDW12575.1 DS232073 EDS33958.1 GECU01001319 JAT06388.1 KQ459603 KPI92724.1 KQ460883 KPJ11232.1 CP012523 ALC38651.1 GFDL01002000 JAV33045.1 GECU01035477 GECU01021350 JAS72229.1 JAS86356.1 CH963920 KRF98791.1 LBMM01000631 KMQ97942.1 OUUW01000006 SPP81567.1 KRF98790.1 EDW78011.2 CH940649 EDW64650.1 AY058354 AAL13583.1 CH477726 EAT36864.1 BABH01005037 CH379062 KRT04988.1 ODYU01003105 SOQ41442.1 BT126166 ADZ99418.1 GFDF01009531 JAV04553.1 CH479184 EDW37304.1 EAL32825.3 AE014134 AAF52449.2 KRT04987.1 GAPW01000535 JAC13063.1 BT031332 AAN10603.1 ABY21745.1 SPP81566.1 SPP81565.1 CM002910 KMY88637.1 KMY88638.1 GFDF01009530 JAV04554.1 CH954177 KQS70650.1 KQS70651.1 EDW64651.1 EDV59141.2 KRF81772.1 CH916372 EDV99191.1 GBXI01002321 JAD11971.1 CH480818 EDW51919.1 GBXI01003929 JAD10363.1 AGBW02008689 OWR52770.1 GFDF01002073 JAV12011.1 GFDF01002075 JAV12009.1 CM000157 KRJ97422.1 EDW87827.2 GFDF01002074 JAV12010.1 DS235123 EEB12185.1 GL436444 EFN71977.1 CH902620 KPU73497.1 GEZM01001569 JAV97664.1 EDV31590.2 NNAY01000372 OXU28817.1 GEZM01001567 JAV97666.1 GEZM01001568 GEZM01001566 JAV97665.1 GEZM01001565 JAV97668.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000069272 UP000075901 UP000000673 UP000075920 UP000075884 UP000075900 UP000075883 UP000076407 UP000075840 UP000075902 UP000007062 UP000075886 UP000075880 UP000075903 UP000076408 UP000075882 UP000030765 UP000075885 UP000235965 UP000053097 UP000279307 UP000095301 UP000009192 UP000002320 UP000092553 UP000091820 UP000007798 UP000036403 UP000268350 UP000008792 UP000008820 UP000001819 UP000008744 UP000000803 UP000008711 UP000001070 UP000001292 UP000002282 UP000192221 UP000095300 UP000009046 UP000000311 UP000007801 UP000215335
UP000069272 UP000075901 UP000000673 UP000075920 UP000075884 UP000075900 UP000075883 UP000076407 UP000075840 UP000075902 UP000007062 UP000075886 UP000075880 UP000075903 UP000076408 UP000075882 UP000030765 UP000075885 UP000235965 UP000053097 UP000279307 UP000095301 UP000009192 UP000002320 UP000092553 UP000091820 UP000007798 UP000036403 UP000268350 UP000008792 UP000008820 UP000001819 UP000008744 UP000000803 UP000008711 UP000001070 UP000001292 UP000002282 UP000192221 UP000095300 UP000009046 UP000000311 UP000007801 UP000215335
Pfam
PF00999 Na_H_Exchanger
Gene 3D
ProteinModelPortal
H9J9L1
A0A2A4K313
A0A2H1WQE8
A0A2W1BUW0
A0A194RD48
A0A194PKB6
+ More
A0A212FG08 A0A0L7LSH2 A0A212FG10 A0A2W1BV43 A0A2A4K4S2 A0A194RGZ3 T1DU38 A0A182FRV9 A0A182T952 W5JHL6 A0A182VRG1 A0A182NFP2 A0A182RX38 A0A182M6L2 A0MNP7 A0A182XGF3 A0A182HZX3 A0A182U2G4 Q7PYT2 A0A182QVG0 A0A182JL69 A0A182UNV4 A0A182XW09 A0A182LR05 A0A084WSH3 A0A182PE22 A0A2J7RJI7 A0A026WQ33 T1PB54 A0A1I8MUY9 A0A1I8MUZ4 A0A2A4JX14 B4KJX2 B0WSL1 A0A1B6K4L7 A0A194PJ27 A0A194R0K4 A0A0M4E945 A0A1Q3FZQ1 A0A1A9WXC6 A0A1B6IHG3 A0A0Q9X3C6 A0A0J7L4S6 A0A3B0JGT3 A0A0Q9X184 B4N076 B4LRP8 Q95U27 Q16R10 H9J9L4 A0A0R3NVL1 A0A2H1VLY0 F2FB79 A0A1L8DDK3 B4GKY8 Q29JY6 Q9VM74 A0A0R3NV36 A0A023EVB6 Q7KTL6 A0A3B0K7E6 A0A3B0JHA4 A0A0J9QX13 A0A0J9QXC9 A0A1L8DE11 A0A0Q5WNB0 A0A0Q5WK11 B4LRP9 B3N5Y2 A0A0Q9WAQ8 B4JQL1 A0A0A1XLT2 B4HXY9 A0A0A1XIM4 A0A212FGB7 A0A1L8E003 A0A1L8E028 A0A0R1DKJ9 B4P000 A0A1W4UNP4 A0A1L8DZT1 A0A1W4UNX2 A0A1I8NMR4 A0A1I8NMR0 E0VFM9 E2A3J5 A0A0P9BNS5 A0A1Y1NIF1 B3MKQ9 A0A232FET6 A0A1Y1NMV7 A0A1Y1NLM9 A0A1Y1NJI0
A0A212FG08 A0A0L7LSH2 A0A212FG10 A0A2W1BV43 A0A2A4K4S2 A0A194RGZ3 T1DU38 A0A182FRV9 A0A182T952 W5JHL6 A0A182VRG1 A0A182NFP2 A0A182RX38 A0A182M6L2 A0MNP7 A0A182XGF3 A0A182HZX3 A0A182U2G4 Q7PYT2 A0A182QVG0 A0A182JL69 A0A182UNV4 A0A182XW09 A0A182LR05 A0A084WSH3 A0A182PE22 A0A2J7RJI7 A0A026WQ33 T1PB54 A0A1I8MUY9 A0A1I8MUZ4 A0A2A4JX14 B4KJX2 B0WSL1 A0A1B6K4L7 A0A194PJ27 A0A194R0K4 A0A0M4E945 A0A1Q3FZQ1 A0A1A9WXC6 A0A1B6IHG3 A0A0Q9X3C6 A0A0J7L4S6 A0A3B0JGT3 A0A0Q9X184 B4N076 B4LRP8 Q95U27 Q16R10 H9J9L4 A0A0R3NVL1 A0A2H1VLY0 F2FB79 A0A1L8DDK3 B4GKY8 Q29JY6 Q9VM74 A0A0R3NV36 A0A023EVB6 Q7KTL6 A0A3B0K7E6 A0A3B0JHA4 A0A0J9QX13 A0A0J9QXC9 A0A1L8DE11 A0A0Q5WNB0 A0A0Q5WK11 B4LRP9 B3N5Y2 A0A0Q9WAQ8 B4JQL1 A0A0A1XLT2 B4HXY9 A0A0A1XIM4 A0A212FGB7 A0A1L8E003 A0A1L8E028 A0A0R1DKJ9 B4P000 A0A1W4UNP4 A0A1L8DZT1 A0A1W4UNX2 A0A1I8NMR4 A0A1I8NMR0 E0VFM9 E2A3J5 A0A0P9BNS5 A0A1Y1NIF1 B3MKQ9 A0A232FET6 A0A1Y1NMV7 A0A1Y1NLM9 A0A1Y1NJI0
Ontologies
GO
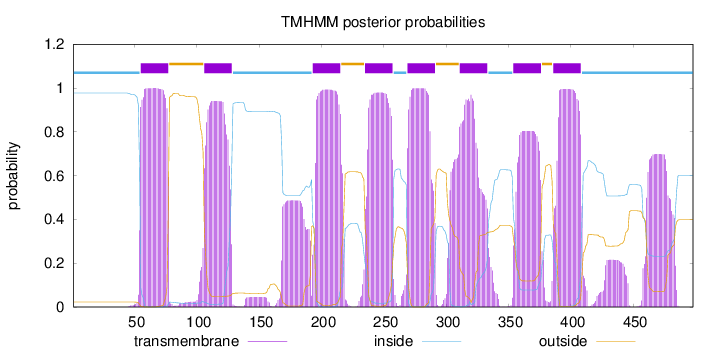
Topology
Length:
498
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
201.40318
Exp number, first 60 AAs:
6.28227
Total prob of N-in:
0.97753
inside
1 - 54
TMhelix
55 - 77
outside
78 - 105
TMhelix
106 - 128
inside
129 - 192
TMhelix
193 - 215
outside
216 - 234
TMhelix
235 - 257
inside
258 - 268
TMhelix
269 - 291
outside
292 - 310
TMhelix
311 - 333
inside
334 - 353
TMhelix
354 - 376
outside
377 - 385
TMhelix
386 - 408
inside
409 - 498
Population Genetic Test Statistics
Pi
284.366635
Theta
168.3732
Tajima's D
2.578256
CLR
0.357341
CSRT
0.949052547372631
Interpretation
Uncertain
