Pre Gene Modal
BGIBMGA006201
Annotation
PREDICTED:_putative_ATP-dependent_RNA_helicase_DHX33_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.784 Nuclear Reliability : 1.188
Sequence
CDS
ATGGATTCAAAGTACTGCTCAATAGGTTTGACCAATGAAACTAAAAATGGTTTTAAGAGGAAGAAACCGACAGCTAGTATTTGTGTGAAAAAGGTCAAATTATTAAGTGATAGTATTAGTAATTGTTTACAAAATGGCCATGAAGTTTCTAAAAACATAGAAACGCATGTTGTTAAAAAAAATTCAGATGATTTGCAAGAAGCTAGAAGGAAACTGCCTGTATTTATGGTTAGAGGCAGGTTATTAGAAGAAATAAGAAAAAACAACACAATGATAATAATAGGTGAGACAGGAAGTGGTAAGACCACTCAGATTCCTCAAATGATTCATGAACAGAGATTAGAAGGCACTGGCTCGATAGCAGTTACTCAGCCTCGTAGAGTAGCTGCTATTACAATTGCATTGCGAGTTGCTGCTGAAATGAATACTGAAGTTGGTAACATTGTTGGGTACTCAGTTAGATTTGAAGATGTGACAAGCCCTAGAACTAAAGTGAAATATTTAACTGATGGTATGTTGCTTAGAGAAGCTATCTTAGATCCTTTGCTCAGTAAATATTCAGTTATTATTCTGGATGAAGCACATGAACGCACTGTGAGCACTGATATATTGTTTGGAATTGTGAAATTAGCACAAAAGGAGAGAAATGAAAAAACTATTAATCCTTTAAAGGTCATTGTAATGTCAGCGACAATGGATGTCGATACATTTAGAAATTATTTTGACAGCTGTCCAGTAATATATTTGGAAGGTCGTACACATCCTGTGACAATATATCATTCTAAAACCAAGCAAGAGGATTACCAGTATGCAGCTGTGTGCACAATATTTCAGCTGCACACTACTACCCCTGCTGATGATGACTTTCTTGTATTTCTAACTGGGCAAGAAGAGATTGAAGCTGTAATGTATAACATAAAACAAATCGCGAAAGAAACAACAGGTCCACCTATACGGGTTTGTCCATTATACGCCGGCCTTCCGCCTGCACAGCAAATGCAGGTGTGGAAAGAGACACCGCCGGGTACCAGGAAAATAGTCCTCGCGACTAATATAGCAGAGGCATCGGTCACGATTCCCAGGATAAAGTGTGTCATTGATACTGGTGTTGTAAAAGAAAGGTCGTGGTGTAGTGCCACAGGTGCGGAGCGGCTGGTGGTGCGCAGCGGGTCGCAGGCGGCGGGCTGGCAGCGCGCCGGCCGCGCGGGCCGCACCGCGCCCGGCGCCGCCTACAGGCTCTATACCGCGCAGCAGTTCAACACAAGACCCTTACATCCTACGCCTGAAATACTTAGATGTCCACTGGCACCAACAATACTGTTGCTAATAGCAGCCGGCGTTGAACCGAGCACATTCCCGCTGATAGACTCTCCACCGCGAGACTCCGTCACTGCTTCATTGACGTTACTTAAGGAATTGGGTGCAATTGACAGCGTAGAAATTCCAAAATTAACCGTATTAGGTCGCAAAATGTCTTCATTCCCGATAGATCCCAAATATTCGAAAGTACTACTGAGTGCCCCCGAATATGGATGCTTAGAAGAGGCTTTAAGTTTAGTAGCAGTTTTGTCTAGTGAAAATATATTCTACACGCCGATGCACAAACGCGAAGAAGCATTGAAAGTAAAGCGAAAATTTATATCGCCAACAGGAGACCACATCACCTTGTTAAATGTTTTTAAAGCATTTTGCAAAGCTACTTTAAAAAAGCAATGGTGTAAAGAAAACTATTTAAATCATCGAAATTTAACGTACGCTTACGAAGTCCGACAGCAGTTACTTGCAATATGTCAGAGATTAAATTTAACTCTATCAAGCTGCGGGTCTGCAACTGATCAGTTGCTGAAGTGTTTGTTGAGCGGCTTATTCAGCAACTGTGCGTGGTCTCGGAGCGCGGGCGTGGCGGGGGGCGGGGCGCGGTACGTGACGGGCGCGGGGGGCGCCGCCAGCCTGCACCCCGCCGCCGCGCTGCACGCCGCCAGCCCGCCCCCCCCCGCGCTGCTCTACACAGAGCTGCTGCACACCCGCCGCACGTACATGATCACCGTCTCCGCCATACAACCGCACTGGCTCCAACAGGTGGCGCCCGATTACGCACGCAGATGTCGGGCTTACCGATGA
Protein
MDSKYCSIGLTNETKNGFKRKKPTASICVKKVKLLSDSISNCLQNGHEVSKNIETHVVKKNSDDLQEARRKLPVFMVRGRLLEEIRKNNTMIIIGETGSGKTTQIPQMIHEQRLEGTGSIAVTQPRRVAAITIALRVAAEMNTEVGNIVGYSVRFEDVTSPRTKVKYLTDGMLLREAILDPLLSKYSVIILDEAHERTVSTDILFGIVKLAQKERNEKTINPLKVIVMSATMDVDTFRNYFDSCPVIYLEGRTHPVTIYHSKTKQEDYQYAAVCTIFQLHTTTPADDDFLVFLTGQEEIEAVMYNIKQIAKETTGPPIRVCPLYAGLPPAQQMQVWKETPPGTRKIVLATNIAEASVTIPRIKCVIDTGVVKERSWCSATGAERLVVRSGSQAAGWQRAGRAGRTAPGAAYRLYTAQQFNTRPLHPTPEILRCPLAPTILLLIAAGVEPSTFPLIDSPPRDSVTASLTLLKELGAIDSVEIPKLTVLGRKMSSFPIDPKYSKVLLSAPEYGCLEEALSLVAVLSSENIFYTPMHKREEALKVKRKFISPTGDHITLLNVFKAFCKATLKKQWCKENYLNHRNLTYAYEVRQQLLAICQRLNLTLSSCGSATDQLLKCLLSGLFSNCAWSRSAGVAGGGARYVTGAGGAASLHPAAALHAASPPPPALLYTELLHTRRTYMITVSAIQPHWLQQVAPDYARRCRAYR
Summary
Uniprot
A0A2H1X0X4
A0A212F0Q2
A0A194R1W1
A0A194PLK8
A0A2P8Y1A3
A0A2J7QBM3
+ More
A0A1B6DZY5 A0A1Y1LTH7 A0A067QN56 E0VC35 D6WNY8 A0A0L7RJV6 A0A1W4WM56 A0A0T6AUJ5 A0A088ATG0 A0A2A3EIW3 A0A1I8MIV4 T1PJ55 N6U2C2 E9GDG1 A0A210Q5A0 A0A195E5N1 F4WP55 A0A026WP55 A0A336MTZ8 A0A0P6ACA7 A0A195B9X2 A0A0C9RDX1 A0A0P6IEX8 A0A195EV48 A0A158NG95 A0A151WMK2 A0A1S3J0N4 W8BBC9 A0A1B0FAS2 A0A0A1X7T3 A0A1L8DGA4 A0A023F2T0 A0A0K8UK92 A0A336MR68 A0A0L0CFP5 A0A151IKT8 A0A1I8NZ77 A0A0A9XAK1 E2A3J9 A0A3B4V1Q6 G3QA12 A0A1L8HGB0 A0A0F8AC25 A0A3P8YYU2 A0A3P8VV43 A0A3B4H2R9 A0A3B3UN32 A0A1S3RN32 A0A2C9JCC8 M3ZU18 A0A0P7V4B5 A0A146NSV0 A0A3B3YKI5 A0A1W4ZAC5 I3KLQ1 H2S6D3 A0A3P8R7H9 A0A3P8R8Q6 A0A3P9D957 A0A087XV01 A0A3B5AGQ2 K7INF0 A0A2U9B9K2 W5L917 A0A3B5L277 A0A2D0QCM4 W5U8G7 A0A3B4AK23 A0A1A9Y702 A0A3P9NRE8 A0A093QIZ5 X1WET4 H2M8J1 F7CJ74 F7CUD1 B1H3C5 A0A182JPN6 A0A3B4E948 A0A2G8KKM0 U5ET14 Q6GL98 A0A0M4EA88 H0Z1C0 A0A1A7Y0P4 A0A182V5S1 A0A182ULL7 B4M9K0 A0A369SQ53 A0A2I4DCN7 A0A182KNZ5 A0A182I1D9 A0A3B3TCG1 H9GLX8
A0A1B6DZY5 A0A1Y1LTH7 A0A067QN56 E0VC35 D6WNY8 A0A0L7RJV6 A0A1W4WM56 A0A0T6AUJ5 A0A088ATG0 A0A2A3EIW3 A0A1I8MIV4 T1PJ55 N6U2C2 E9GDG1 A0A210Q5A0 A0A195E5N1 F4WP55 A0A026WP55 A0A336MTZ8 A0A0P6ACA7 A0A195B9X2 A0A0C9RDX1 A0A0P6IEX8 A0A195EV48 A0A158NG95 A0A151WMK2 A0A1S3J0N4 W8BBC9 A0A1B0FAS2 A0A0A1X7T3 A0A1L8DGA4 A0A023F2T0 A0A0K8UK92 A0A336MR68 A0A0L0CFP5 A0A151IKT8 A0A1I8NZ77 A0A0A9XAK1 E2A3J9 A0A3B4V1Q6 G3QA12 A0A1L8HGB0 A0A0F8AC25 A0A3P8YYU2 A0A3P8VV43 A0A3B4H2R9 A0A3B3UN32 A0A1S3RN32 A0A2C9JCC8 M3ZU18 A0A0P7V4B5 A0A146NSV0 A0A3B3YKI5 A0A1W4ZAC5 I3KLQ1 H2S6D3 A0A3P8R7H9 A0A3P8R8Q6 A0A3P9D957 A0A087XV01 A0A3B5AGQ2 K7INF0 A0A2U9B9K2 W5L917 A0A3B5L277 A0A2D0QCM4 W5U8G7 A0A3B4AK23 A0A1A9Y702 A0A3P9NRE8 A0A093QIZ5 X1WET4 H2M8J1 F7CJ74 F7CUD1 B1H3C5 A0A182JPN6 A0A3B4E948 A0A2G8KKM0 U5ET14 Q6GL98 A0A0M4EA88 H0Z1C0 A0A1A7Y0P4 A0A182V5S1 A0A182ULL7 B4M9K0 A0A369SQ53 A0A2I4DCN7 A0A182KNZ5 A0A182I1D9 A0A3B3TCG1 H9GLX8
Pubmed
22118469
26354079
29403074
28004739
24845553
20566863
+ More
18362917 19820115 25315136 23537049 21292972 28812685 21719571 24508170 30249741 21347285 24495485 25830018 25474469 26108605 25401762 20798317 27762356 25835551 25069045 24487278 15562597 23542700 25186727 21551351 20075255 25329095 23127152 25463417 23594743 17554307 20431018 29023486 20360741 17994087 18057021 30042472 20966253 29240929 21881562
18362917 19820115 25315136 23537049 21292972 28812685 21719571 24508170 30249741 21347285 24495485 25830018 25474469 26108605 25401762 20798317 27762356 25835551 25069045 24487278 15562597 23542700 25186727 21551351 20075255 25329095 23127152 25463417 23594743 17554307 20431018 29023486 20360741 17994087 18057021 30042472 20966253 29240929 21881562
EMBL
ODYU01012586
SOQ58995.1
AGBW02011047
OWR47310.1
KQ460870
KPJ11773.1
+ More
KQ459601 KPI93883.1 PYGN01001054 PSN38038.1 NEVH01016296 PNF25969.1 GEDC01006077 JAS31221.1 GEZM01048895 JAV76301.1 KK853138 KDR10772.1 DS235045 EEB10941.1 KQ971343 EFA03189.1 KQ414578 KOC71150.1 LJIG01022848 KRT78407.1 KZ288253 PBC30961.1 KA648719 AFP63348.1 APGK01045200 APGK01045201 APGK01045202 APGK01045203 KB741031 ENN74771.1 GL732540 EFX82076.1 NEDP02004983 OWF43914.1 KQ979608 KYN20396.1 GL888243 EGI63986.1 KK107139 QOIP01000003 EZA57800.1 RLU24693.1 UFQT01002065 SSX32569.1 GDIP01031344 JAM72371.1 KQ976542 KYM81030.1 GBYB01011282 JAG81049.1 GDIQ01007081 JAN87656.1 KQ981965 KYN31767.1 ADTU01015078 ADTU01015079 KQ982944 KYQ48935.1 GAMC01008020 JAB98535.1 CCAG010015740 GBXI01007326 JAD06966.1 GFDF01008667 JAV05417.1 GBBI01003214 JAC15498.1 GDHF01025584 JAI26730.1 SSX32570.1 JRES01000453 KNC31065.1 KQ977164 KYN05222.1 GBHO01025847 JAG17757.1 GL436444 EFN71981.1 CM004468 OCT95132.1 KQ042784 KKF10644.1 JARO02004955 KPP67604.1 GCES01151787 JAQ34535.1 AERX01048964 AYCK01007202 CP026246 AWP00442.1 JT408184 AHH38261.1 KL672574 KFW86370.1 CT573103 AAMC01010900 BC161345 AAI61345.1 MRZV01000516 PIK48549.1 GANO01002973 JAB56898.1 BC074605 AAH74605.1 CP012523 ALC40215.1 ABQF01026134 HADW01017903 HADX01001659 SBP23891.1 CH940654 EDW57876.1 KRF77997.1 NOWV01000002 RDD47573.1 APCN01000054 AAWZ02026898 AAWZ02026899 AAWZ02026900 AAWZ02026901 AAWZ02026902
KQ459601 KPI93883.1 PYGN01001054 PSN38038.1 NEVH01016296 PNF25969.1 GEDC01006077 JAS31221.1 GEZM01048895 JAV76301.1 KK853138 KDR10772.1 DS235045 EEB10941.1 KQ971343 EFA03189.1 KQ414578 KOC71150.1 LJIG01022848 KRT78407.1 KZ288253 PBC30961.1 KA648719 AFP63348.1 APGK01045200 APGK01045201 APGK01045202 APGK01045203 KB741031 ENN74771.1 GL732540 EFX82076.1 NEDP02004983 OWF43914.1 KQ979608 KYN20396.1 GL888243 EGI63986.1 KK107139 QOIP01000003 EZA57800.1 RLU24693.1 UFQT01002065 SSX32569.1 GDIP01031344 JAM72371.1 KQ976542 KYM81030.1 GBYB01011282 JAG81049.1 GDIQ01007081 JAN87656.1 KQ981965 KYN31767.1 ADTU01015078 ADTU01015079 KQ982944 KYQ48935.1 GAMC01008020 JAB98535.1 CCAG010015740 GBXI01007326 JAD06966.1 GFDF01008667 JAV05417.1 GBBI01003214 JAC15498.1 GDHF01025584 JAI26730.1 SSX32570.1 JRES01000453 KNC31065.1 KQ977164 KYN05222.1 GBHO01025847 JAG17757.1 GL436444 EFN71981.1 CM004468 OCT95132.1 KQ042784 KKF10644.1 JARO02004955 KPP67604.1 GCES01151787 JAQ34535.1 AERX01048964 AYCK01007202 CP026246 AWP00442.1 JT408184 AHH38261.1 KL672574 KFW86370.1 CT573103 AAMC01010900 BC161345 AAI61345.1 MRZV01000516 PIK48549.1 GANO01002973 JAB56898.1 BC074605 AAH74605.1 CP012523 ALC40215.1 ABQF01026134 HADW01017903 HADX01001659 SBP23891.1 CH940654 EDW57876.1 KRF77997.1 NOWV01000002 RDD47573.1 APCN01000054 AAWZ02026898 AAWZ02026899 AAWZ02026900 AAWZ02026901 AAWZ02026902
Proteomes
UP000007151
UP000053240
UP000053268
UP000245037
UP000235965
UP000027135
+ More
UP000009046 UP000007266 UP000053825 UP000192223 UP000005203 UP000242457 UP000095301 UP000019118 UP000000305 UP000242188 UP000078492 UP000007755 UP000053097 UP000279307 UP000078540 UP000078541 UP000005205 UP000075809 UP000085678 UP000092444 UP000037069 UP000078542 UP000095300 UP000000311 UP000261420 UP000007635 UP000186698 UP000265140 UP000265120 UP000261460 UP000261500 UP000087266 UP000076420 UP000002852 UP000034805 UP000261480 UP000192224 UP000005207 UP000005226 UP000265100 UP000265160 UP000028760 UP000261400 UP000002358 UP000246464 UP000018467 UP000261380 UP000221080 UP000261520 UP000092443 UP000242638 UP000053258 UP000000437 UP000001038 UP000008143 UP000075881 UP000261440 UP000230750 UP000092553 UP000007754 UP000075903 UP000075902 UP000008792 UP000253843 UP000192220 UP000075882 UP000075840 UP000261540 UP000001646
UP000009046 UP000007266 UP000053825 UP000192223 UP000005203 UP000242457 UP000095301 UP000019118 UP000000305 UP000242188 UP000078492 UP000007755 UP000053097 UP000279307 UP000078540 UP000078541 UP000005205 UP000075809 UP000085678 UP000092444 UP000037069 UP000078542 UP000095300 UP000000311 UP000261420 UP000007635 UP000186698 UP000265140 UP000265120 UP000261460 UP000261500 UP000087266 UP000076420 UP000002852 UP000034805 UP000261480 UP000192224 UP000005207 UP000005226 UP000265100 UP000265160 UP000028760 UP000261400 UP000002358 UP000246464 UP000018467 UP000261380 UP000221080 UP000261520 UP000092443 UP000242638 UP000053258 UP000000437 UP000001038 UP000008143 UP000075881 UP000261440 UP000230750 UP000092553 UP000007754 UP000075903 UP000075902 UP000008792 UP000253843 UP000192220 UP000075882 UP000075840 UP000261540 UP000001646
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
A0A2H1X0X4
A0A212F0Q2
A0A194R1W1
A0A194PLK8
A0A2P8Y1A3
A0A2J7QBM3
+ More
A0A1B6DZY5 A0A1Y1LTH7 A0A067QN56 E0VC35 D6WNY8 A0A0L7RJV6 A0A1W4WM56 A0A0T6AUJ5 A0A088ATG0 A0A2A3EIW3 A0A1I8MIV4 T1PJ55 N6U2C2 E9GDG1 A0A210Q5A0 A0A195E5N1 F4WP55 A0A026WP55 A0A336MTZ8 A0A0P6ACA7 A0A195B9X2 A0A0C9RDX1 A0A0P6IEX8 A0A195EV48 A0A158NG95 A0A151WMK2 A0A1S3J0N4 W8BBC9 A0A1B0FAS2 A0A0A1X7T3 A0A1L8DGA4 A0A023F2T0 A0A0K8UK92 A0A336MR68 A0A0L0CFP5 A0A151IKT8 A0A1I8NZ77 A0A0A9XAK1 E2A3J9 A0A3B4V1Q6 G3QA12 A0A1L8HGB0 A0A0F8AC25 A0A3P8YYU2 A0A3P8VV43 A0A3B4H2R9 A0A3B3UN32 A0A1S3RN32 A0A2C9JCC8 M3ZU18 A0A0P7V4B5 A0A146NSV0 A0A3B3YKI5 A0A1W4ZAC5 I3KLQ1 H2S6D3 A0A3P8R7H9 A0A3P8R8Q6 A0A3P9D957 A0A087XV01 A0A3B5AGQ2 K7INF0 A0A2U9B9K2 W5L917 A0A3B5L277 A0A2D0QCM4 W5U8G7 A0A3B4AK23 A0A1A9Y702 A0A3P9NRE8 A0A093QIZ5 X1WET4 H2M8J1 F7CJ74 F7CUD1 B1H3C5 A0A182JPN6 A0A3B4E948 A0A2G8KKM0 U5ET14 Q6GL98 A0A0M4EA88 H0Z1C0 A0A1A7Y0P4 A0A182V5S1 A0A182ULL7 B4M9K0 A0A369SQ53 A0A2I4DCN7 A0A182KNZ5 A0A182I1D9 A0A3B3TCG1 H9GLX8
A0A1B6DZY5 A0A1Y1LTH7 A0A067QN56 E0VC35 D6WNY8 A0A0L7RJV6 A0A1W4WM56 A0A0T6AUJ5 A0A088ATG0 A0A2A3EIW3 A0A1I8MIV4 T1PJ55 N6U2C2 E9GDG1 A0A210Q5A0 A0A195E5N1 F4WP55 A0A026WP55 A0A336MTZ8 A0A0P6ACA7 A0A195B9X2 A0A0C9RDX1 A0A0P6IEX8 A0A195EV48 A0A158NG95 A0A151WMK2 A0A1S3J0N4 W8BBC9 A0A1B0FAS2 A0A0A1X7T3 A0A1L8DGA4 A0A023F2T0 A0A0K8UK92 A0A336MR68 A0A0L0CFP5 A0A151IKT8 A0A1I8NZ77 A0A0A9XAK1 E2A3J9 A0A3B4V1Q6 G3QA12 A0A1L8HGB0 A0A0F8AC25 A0A3P8YYU2 A0A3P8VV43 A0A3B4H2R9 A0A3B3UN32 A0A1S3RN32 A0A2C9JCC8 M3ZU18 A0A0P7V4B5 A0A146NSV0 A0A3B3YKI5 A0A1W4ZAC5 I3KLQ1 H2S6D3 A0A3P8R7H9 A0A3P8R8Q6 A0A3P9D957 A0A087XV01 A0A3B5AGQ2 K7INF0 A0A2U9B9K2 W5L917 A0A3B5L277 A0A2D0QCM4 W5U8G7 A0A3B4AK23 A0A1A9Y702 A0A3P9NRE8 A0A093QIZ5 X1WET4 H2M8J1 F7CJ74 F7CUD1 B1H3C5 A0A182JPN6 A0A3B4E948 A0A2G8KKM0 U5ET14 Q6GL98 A0A0M4EA88 H0Z1C0 A0A1A7Y0P4 A0A182V5S1 A0A182ULL7 B4M9K0 A0A369SQ53 A0A2I4DCN7 A0A182KNZ5 A0A182I1D9 A0A3B3TCG1 H9GLX8
PDB
6I3P
E-value=4.87917e-125,
Score=1149
Ontologies
GO
Topology
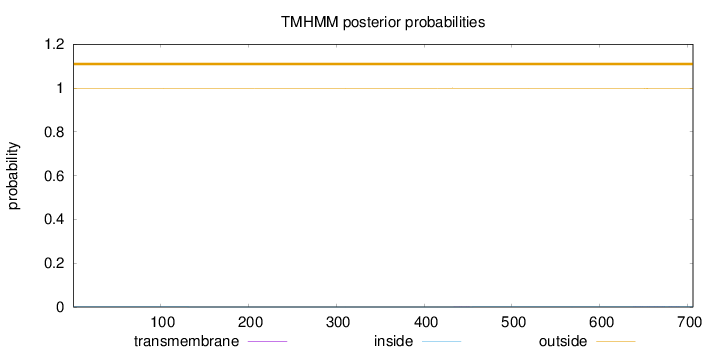
Length:
706
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03209
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00059
outside
1 - 706
Population Genetic Test Statistics
Pi
272.871837
Theta
203.460477
Tajima's D
1.484614
CLR
0.265598
CSRT
0.784010799460027
Interpretation
Possibly Positive selection
