Pre Gene Modal
BGIBMGA005943
Annotation
PREDICTED:_FAD-dependent_oxidoreductase_domain-containing_protein_1_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.246 Nuclear Reliability : 2.967 PlasmaMembrane Reliability : 1.667
Sequence
CDS
ATGATAAAAACTTGGAATATAATGACGAAATCTAATACAAGATTTTCGTACCCAGAAAATACTGATATTGTTATAATAGGTGGTGGTATAATAGGAGCAGCAACAGCTTATTGGTTAAAGCGCAGAGCTGGTGATGGTCTCTCAGTAGTGGTCATAGAAAAAGATTTTTCCTATAAACAAGCTCAAAGGCACCTTCAGCATGGAACACTTTCACAACATTTCACACTACCAGAAAATTTGTTCCTATCGCAATTCAGTGCATTCTTCCTTAGAAACATTAGAGAACATCTTGGTGAAGATATAAATGTTGAATATTGTCCAACTGGGTCATTAGTACTGGCGAGTAACGACTATGCACAAAAATTAGAAGAAACTTCTTCATTATTGAAGGAACATGGGTCACCAAATGAACTTTTGTCTCCTAAGGAAATAAATAAAAAGTTTCCATGGATAAATACTGCAGATATCAAGTTAGGATGCATGGGTGTAGAATCTGAGGGAACATTTAATGGATGGGAACTACATAAAGGCTTTGTAAAAAAGTCAAAAGAACTTGGAGCATTATATATTAATGCTGAAGTAGTTGGTTTTGATTTGGAGCAACAGCGAGATGTTCTAATGGAAGGTGTCACCCCAGGTGCATTTGAAAGGATAATGCGAGTTAGATACATGACAGAAGAAAATGAAGAATATGGAATAAAATTTGCTGGATGCATATTAACAGCTGGTTCTGATTCTAGTGAAATAGCTAAATTTGCAAAAGTAGGTACTGGTGATGGAATGCTCATGGTACCATTACCTGTTGAGAAAAGAAAACATGAAGTTTATTCTCTTGAAGGAAATAATACAGAAATTGGTCTTAATACTCCCATGATAATGGATACAACTGGGTTATGGTTACAAAGAAAAGGTTTAGGAAAGGACTTCTTAGGTGGATATTTACCTTCACTAGATCAAAATGATAATCTTGTTGATACAGAACAGTATTTGAAAACAACTTTTCTATCATCATTAATGCATAGAATCCCTAGCTGTAATAATGCTCAGGTACAAAGCCATAGCAGTTTGTATTATGATTGCACCACATACGACAATTCAGGAATATTGGGACCACATCCTTATCATAATAATTTGTATATTGCTGCAGGATTTGGTCAGCTAGGTTGTCAACATGCTCCGGGTTTGGGAAGAGCTCTAACGGAACTAATCATAGACAGTCAGTTCAATACTATCGACTTGACAAGGCTTGGATTTGACAGATTATTAACTGATCAACCTATGATGGAATGCAATGCTTATTGA
Protein
MIKTWNIMTKSNTRFSYPENTDIVIIGGGIIGAATAYWLKRRAGDGLSVVVIEKDFSYKQAQRHLQHGTLSQHFTLPENLFLSQFSAFFLRNIREHLGEDINVEYCPTGSLVLASNDYAQKLEETSSLLKEHGSPNELLSPKEINKKFPWINTADIKLGCMGVESEGTFNGWELHKGFVKKSKELGALYINAEVVGFDLEQQRDVLMEGVTPGAFERIMRVRYMTEENEEYGIKFAGCILTAGSDSSEIAKFAKVGTGDGMLMVPLPVEKRKHEVYSLEGNNTEIGLNTPMIMDTTGLWLQRKGLGKDFLGGYLPSLDQNDNLVDTEQYLKTTFLSSLMHRIPSCNNAQVQSHSSLYYDCTTYDNSGILGPHPYHNNLYIAAGFGQLGCQHAPGLGRALTELIIDSQFNTIDLTRLGFDRLLTDQPMMECNAY
Summary
Uniprot
H9J8V1
A0A194PMC2
A0A194R250
A0A2A4IXJ2
A0A2W1BZ34
A0A212F0T4
+ More
S4PPD5 A0A2H1X2X9 A0A1L8DAR2 A0A1L8DAP0 A0A182R9S2 A0A084VMC5 B0WCQ5 A0A182LU44 Q16HC9 A0A182H861 Q16N12 A0A182IKV2 A0A182WGX2 A0A2J7Q5Q8 A0A1I8N1X4 T1PF41 A0A182QTS3 A0A2J7Q5P7 A0A182YFG1 A0A182T3I5 A0A1W4WG11 Q7PW61 A0A1I8NZ45 A0A2M4BJZ3 A0A2M4BJX5 A0A067RS91 A0A1S4H017 A0A182KUS6 A0A182I4X0 A0A182X2Y4 A0A182ULX1 U5EGT3 A0A182NEV9 A0A2J7Q5P8 W5J8I6 A0A182UG59 A0A1Y1LDJ0 A0A182K3Q1 A0A2M4AG96 A0A2J7Q5P0 A0A1Q3FJ94 A0A1B0A538 A0A2M4AG02 A0A1A9W064 A0A182PDF3 D3TRX6 B4LS15 A0A0M4ERA7 A0A0L0BQE6 A0A0C9PVC0 B4KE65 A0A1B0B7N6 A0A1A9XL17 D6WK94 A0A1A9V9I0 A0A182F5I7 B4N0X8 A0A0L7KY90 Q9VJ10 A0A2J7Q5P6 T1GPK0 A0A3B0KKA3 Q29PI3 B4I5Q4 B4GKT6 B4Q9F3 N6TM66 B3NMA3 A0A0A1X617 J3JYT6 W8BW46 A0A1W4VXU0 A0A1J1I3D8 B4PAF9 A0A0A1XB41 A0A0K8UR91 B3MNQ8 A0A2J7Q5P9 A0A034VRX3 A0A034W0E1 A0A034W2Y0 A0A0M3QVD2 W8C9U3 A0A0K8VT08 K7IRF3 B4KNI5 B3MIP7 A0A1B6F1W8 A0A2M3ZIC9 B4J6J4 B4LK08 A0A195F9S8 A0A2M3Z9K5 T1I709
S4PPD5 A0A2H1X2X9 A0A1L8DAR2 A0A1L8DAP0 A0A182R9S2 A0A084VMC5 B0WCQ5 A0A182LU44 Q16HC9 A0A182H861 Q16N12 A0A182IKV2 A0A182WGX2 A0A2J7Q5Q8 A0A1I8N1X4 T1PF41 A0A182QTS3 A0A2J7Q5P7 A0A182YFG1 A0A182T3I5 A0A1W4WG11 Q7PW61 A0A1I8NZ45 A0A2M4BJZ3 A0A2M4BJX5 A0A067RS91 A0A1S4H017 A0A182KUS6 A0A182I4X0 A0A182X2Y4 A0A182ULX1 U5EGT3 A0A182NEV9 A0A2J7Q5P8 W5J8I6 A0A182UG59 A0A1Y1LDJ0 A0A182K3Q1 A0A2M4AG96 A0A2J7Q5P0 A0A1Q3FJ94 A0A1B0A538 A0A2M4AG02 A0A1A9W064 A0A182PDF3 D3TRX6 B4LS15 A0A0M4ERA7 A0A0L0BQE6 A0A0C9PVC0 B4KE65 A0A1B0B7N6 A0A1A9XL17 D6WK94 A0A1A9V9I0 A0A182F5I7 B4N0X8 A0A0L7KY90 Q9VJ10 A0A2J7Q5P6 T1GPK0 A0A3B0KKA3 Q29PI3 B4I5Q4 B4GKT6 B4Q9F3 N6TM66 B3NMA3 A0A0A1X617 J3JYT6 W8BW46 A0A1W4VXU0 A0A1J1I3D8 B4PAF9 A0A0A1XB41 A0A0K8UR91 B3MNQ8 A0A2J7Q5P9 A0A034VRX3 A0A034W0E1 A0A034W2Y0 A0A0M3QVD2 W8C9U3 A0A0K8VT08 K7IRF3 B4KNI5 B3MIP7 A0A1B6F1W8 A0A2M3ZIC9 B4J6J4 B4LK08 A0A195F9S8 A0A2M3Z9K5 T1I709
Pubmed
19121390
26354079
28756777
22118469
23622113
24438588
+ More
17510324 26483478 25315136 25244985 12364791 24845553 20966253 20920257 23761445 28004739 20353571 17994087 26108605 18057021 18362917 19820115 26227816 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249 23537049 25830018 22516182 24495485 17550304 25348373 20075255
17510324 26483478 25315136 25244985 12364791 24845553 20966253 20920257 23761445 28004739 20353571 17994087 26108605 18057021 18362917 19820115 26227816 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 22936249 23537049 25830018 22516182 24495485 17550304 25348373 20075255
EMBL
BABH01005005
BABH01005006
KQ459601
KPI93884.1
KQ460870
KPJ11772.1
+ More
NWSH01005932 PCG63833.1 KZ149895 PZC78844.1 AGBW02011047 OWR47311.1 GAIX01000312 JAA92248.1 ODYU01012586 SOQ58994.1 GFDF01010528 JAV03556.1 GFDF01010556 JAV03528.1 ATLV01014585 KE524975 KFB39119.1 DS231890 EDS43781.1 AXCM01001875 CH478183 EAT33646.1 JXUM01028759 KQ560831 KXJ80698.1 CH477842 EAT35741.1 NEVH01017553 PNF23905.1 KA647309 AFP61938.1 AXCN02002830 PNF23904.1 AAAB01008984 EAA14740.4 GGFJ01004234 MBW53375.1 GGFJ01004235 MBW53376.1 KK852451 KDR23675.1 APCN01003737 GANO01003313 JAB56558.1 PNF23907.1 ADMH02001866 ETN60772.1 GEZM01060706 JAV70951.1 GGFK01006490 MBW39811.1 PNF23903.1 GFDL01007451 JAV27594.1 GGFK01006404 MBW39725.1 CCAG010004816 EZ424178 ADD20454.1 CH940649 EDW63691.1 CP012523 ALC39675.1 JRES01001517 KNC22310.1 GBYB01005368 JAG75135.1 CH933807 EDW11810.2 KRG02923.1 JXJN01009696 KQ971342 EFA03961.2 CH963920 EDW78140.1 JTDY01004557 KOB68014.1 AE014134 AY113419 AAF53748.1 AAM29424.1 PNF23906.1 CAQQ02036668 OUUW01000010 SPP86216.1 CH379058 EAL34310.1 CH480822 EDW55710.1 CH479184 EDW37252.1 CM000361 CM002910 EDX05460.1 KMY90931.1 APGK01018911 KB740085 ENN81544.1 CH954179 EDV54703.1 GBXI01009090 GBXI01007705 JAD05202.1 JAD06587.1 BT128417 AEE63374.1 GAMC01009074 JAB97481.1 CVRI01000038 CRK94290.1 CM000158 EDW90367.1 GBXI01006096 JAD08196.1 GDHF01023110 JAI29204.1 CH902620 EDV31145.1 PNF23908.1 GAKP01012895 GAKP01012891 JAC46061.1 GAKP01009892 GAKP01009891 JAC49061.1 GAKP01009893 GAKP01009890 JAC49059.1 CP012524 ALC42173.1 GAMC01001854 JAC04702.1 GDHF01010331 JAI41983.1 CH933808 EDW08944.1 CH902619 EDV38123.1 GECZ01025622 JAS44147.1 GGFM01007518 MBW28269.1 CH916367 EDW01995.1 CH940648 EDW61662.1 KQ981727 KYN36804.1 GGFM01004367 MBW25118.1 ACPB03005126
NWSH01005932 PCG63833.1 KZ149895 PZC78844.1 AGBW02011047 OWR47311.1 GAIX01000312 JAA92248.1 ODYU01012586 SOQ58994.1 GFDF01010528 JAV03556.1 GFDF01010556 JAV03528.1 ATLV01014585 KE524975 KFB39119.1 DS231890 EDS43781.1 AXCM01001875 CH478183 EAT33646.1 JXUM01028759 KQ560831 KXJ80698.1 CH477842 EAT35741.1 NEVH01017553 PNF23905.1 KA647309 AFP61938.1 AXCN02002830 PNF23904.1 AAAB01008984 EAA14740.4 GGFJ01004234 MBW53375.1 GGFJ01004235 MBW53376.1 KK852451 KDR23675.1 APCN01003737 GANO01003313 JAB56558.1 PNF23907.1 ADMH02001866 ETN60772.1 GEZM01060706 JAV70951.1 GGFK01006490 MBW39811.1 PNF23903.1 GFDL01007451 JAV27594.1 GGFK01006404 MBW39725.1 CCAG010004816 EZ424178 ADD20454.1 CH940649 EDW63691.1 CP012523 ALC39675.1 JRES01001517 KNC22310.1 GBYB01005368 JAG75135.1 CH933807 EDW11810.2 KRG02923.1 JXJN01009696 KQ971342 EFA03961.2 CH963920 EDW78140.1 JTDY01004557 KOB68014.1 AE014134 AY113419 AAF53748.1 AAM29424.1 PNF23906.1 CAQQ02036668 OUUW01000010 SPP86216.1 CH379058 EAL34310.1 CH480822 EDW55710.1 CH479184 EDW37252.1 CM000361 CM002910 EDX05460.1 KMY90931.1 APGK01018911 KB740085 ENN81544.1 CH954179 EDV54703.1 GBXI01009090 GBXI01007705 JAD05202.1 JAD06587.1 BT128417 AEE63374.1 GAMC01009074 JAB97481.1 CVRI01000038 CRK94290.1 CM000158 EDW90367.1 GBXI01006096 JAD08196.1 GDHF01023110 JAI29204.1 CH902620 EDV31145.1 PNF23908.1 GAKP01012895 GAKP01012891 JAC46061.1 GAKP01009892 GAKP01009891 JAC49061.1 GAKP01009893 GAKP01009890 JAC49059.1 CP012524 ALC42173.1 GAMC01001854 JAC04702.1 GDHF01010331 JAI41983.1 CH933808 EDW08944.1 CH902619 EDV38123.1 GECZ01025622 JAS44147.1 GGFM01007518 MBW28269.1 CH916367 EDW01995.1 CH940648 EDW61662.1 KQ981727 KYN36804.1 GGFM01004367 MBW25118.1 ACPB03005126
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000075900
+ More
UP000030765 UP000002320 UP000075883 UP000008820 UP000069940 UP000249989 UP000075880 UP000075920 UP000235965 UP000095301 UP000075886 UP000076408 UP000075901 UP000192223 UP000007062 UP000095300 UP000027135 UP000075882 UP000075840 UP000076407 UP000075903 UP000075884 UP000000673 UP000075902 UP000075881 UP000092445 UP000091820 UP000075885 UP000092444 UP000008792 UP000092553 UP000037069 UP000009192 UP000092460 UP000092443 UP000007266 UP000078200 UP000069272 UP000007798 UP000037510 UP000000803 UP000015102 UP000268350 UP000001819 UP000001292 UP000008744 UP000000304 UP000019118 UP000008711 UP000192221 UP000183832 UP000002282 UP000007801 UP000002358 UP000001070 UP000078541 UP000015103
UP000030765 UP000002320 UP000075883 UP000008820 UP000069940 UP000249989 UP000075880 UP000075920 UP000235965 UP000095301 UP000075886 UP000076408 UP000075901 UP000192223 UP000007062 UP000095300 UP000027135 UP000075882 UP000075840 UP000076407 UP000075903 UP000075884 UP000000673 UP000075902 UP000075881 UP000092445 UP000091820 UP000075885 UP000092444 UP000008792 UP000092553 UP000037069 UP000009192 UP000092460 UP000092443 UP000007266 UP000078200 UP000069272 UP000007798 UP000037510 UP000000803 UP000015102 UP000268350 UP000001819 UP000001292 UP000008744 UP000000304 UP000019118 UP000008711 UP000192221 UP000183832 UP000002282 UP000007801 UP000002358 UP000001070 UP000078541 UP000015103
Interpro
Gene 3D
ProteinModelPortal
H9J8V1
A0A194PMC2
A0A194R250
A0A2A4IXJ2
A0A2W1BZ34
A0A212F0T4
+ More
S4PPD5 A0A2H1X2X9 A0A1L8DAR2 A0A1L8DAP0 A0A182R9S2 A0A084VMC5 B0WCQ5 A0A182LU44 Q16HC9 A0A182H861 Q16N12 A0A182IKV2 A0A182WGX2 A0A2J7Q5Q8 A0A1I8N1X4 T1PF41 A0A182QTS3 A0A2J7Q5P7 A0A182YFG1 A0A182T3I5 A0A1W4WG11 Q7PW61 A0A1I8NZ45 A0A2M4BJZ3 A0A2M4BJX5 A0A067RS91 A0A1S4H017 A0A182KUS6 A0A182I4X0 A0A182X2Y4 A0A182ULX1 U5EGT3 A0A182NEV9 A0A2J7Q5P8 W5J8I6 A0A182UG59 A0A1Y1LDJ0 A0A182K3Q1 A0A2M4AG96 A0A2J7Q5P0 A0A1Q3FJ94 A0A1B0A538 A0A2M4AG02 A0A1A9W064 A0A182PDF3 D3TRX6 B4LS15 A0A0M4ERA7 A0A0L0BQE6 A0A0C9PVC0 B4KE65 A0A1B0B7N6 A0A1A9XL17 D6WK94 A0A1A9V9I0 A0A182F5I7 B4N0X8 A0A0L7KY90 Q9VJ10 A0A2J7Q5P6 T1GPK0 A0A3B0KKA3 Q29PI3 B4I5Q4 B4GKT6 B4Q9F3 N6TM66 B3NMA3 A0A0A1X617 J3JYT6 W8BW46 A0A1W4VXU0 A0A1J1I3D8 B4PAF9 A0A0A1XB41 A0A0K8UR91 B3MNQ8 A0A2J7Q5P9 A0A034VRX3 A0A034W0E1 A0A034W2Y0 A0A0M3QVD2 W8C9U3 A0A0K8VT08 K7IRF3 B4KNI5 B3MIP7 A0A1B6F1W8 A0A2M3ZIC9 B4J6J4 B4LK08 A0A195F9S8 A0A2M3Z9K5 T1I709
S4PPD5 A0A2H1X2X9 A0A1L8DAR2 A0A1L8DAP0 A0A182R9S2 A0A084VMC5 B0WCQ5 A0A182LU44 Q16HC9 A0A182H861 Q16N12 A0A182IKV2 A0A182WGX2 A0A2J7Q5Q8 A0A1I8N1X4 T1PF41 A0A182QTS3 A0A2J7Q5P7 A0A182YFG1 A0A182T3I5 A0A1W4WG11 Q7PW61 A0A1I8NZ45 A0A2M4BJZ3 A0A2M4BJX5 A0A067RS91 A0A1S4H017 A0A182KUS6 A0A182I4X0 A0A182X2Y4 A0A182ULX1 U5EGT3 A0A182NEV9 A0A2J7Q5P8 W5J8I6 A0A182UG59 A0A1Y1LDJ0 A0A182K3Q1 A0A2M4AG96 A0A2J7Q5P0 A0A1Q3FJ94 A0A1B0A538 A0A2M4AG02 A0A1A9W064 A0A182PDF3 D3TRX6 B4LS15 A0A0M4ERA7 A0A0L0BQE6 A0A0C9PVC0 B4KE65 A0A1B0B7N6 A0A1A9XL17 D6WK94 A0A1A9V9I0 A0A182F5I7 B4N0X8 A0A0L7KY90 Q9VJ10 A0A2J7Q5P6 T1GPK0 A0A3B0KKA3 Q29PI3 B4I5Q4 B4GKT6 B4Q9F3 N6TM66 B3NMA3 A0A0A1X617 J3JYT6 W8BW46 A0A1W4VXU0 A0A1J1I3D8 B4PAF9 A0A0A1XB41 A0A0K8UR91 B3MNQ8 A0A2J7Q5P9 A0A034VRX3 A0A034W0E1 A0A034W2Y0 A0A0M3QVD2 W8C9U3 A0A0K8VT08 K7IRF3 B4KNI5 B3MIP7 A0A1B6F1W8 A0A2M3ZIC9 B4J6J4 B4LK08 A0A195F9S8 A0A2M3Z9K5 T1I709
PDB
1Y56
E-value=1.15064e-05,
Score=117
Ontologies
GO
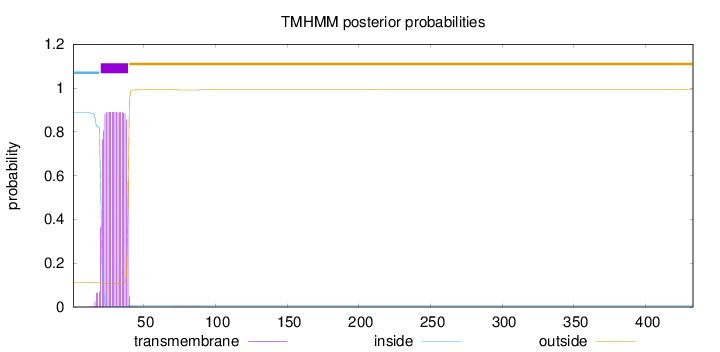
Topology
Length:
433
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
16.98302
Exp number, first 60 AAs:
16.97781
Total prob of N-in:
0.88852
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 39
outside
40 - 433
Population Genetic Test Statistics
Pi
241.102236
Theta
173.66334
Tajima's D
0.27653
CLR
0.330032
CSRT
0.450677466126694
Interpretation
Uncertain
