Gene
KWMTBOMO01892
Pre Gene Modal
BGIBMGA006199
Annotation
PREDICTED:_uncharacterized_protein_LOC101738263_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.701 PlasmaMembrane Reliability : 4.962
Sequence
CDS
ATGAATAATGCGGATAACAGCATTGAGTTTTCTGAAGTCAATGAAAGAAATGGAAACGTACACGAAGAAAAAATACGCAAAGGTTGGGTACAATTTGATGACGAACAACAAACACCAACTGTTTCCCATTCTGAAAATGAAATAGCAGTAACAAAAACACCACCTGTTGCTTCAACGCAAACCCCGACACGTCCTACTGTTCCCGCTGTTTTGAATACAGAAACAGTTCACGTGAATCTTGAAAGAGGAGATAAAAATTTGGAATCCAGTACTCAATCAACTTTAACGAAAAATGTTGAATTCGTCAACGTACGCCATGGGTTCTCTAATGGTGACATAATAGTGACATTGTTACCTGTTAACACTAAGTGGCCGTGGATTACAGCAGCTCAGTTTAGGCCAGAACTAGTACCTGAAGAATTAATGGCACAAGGCTTAACTTTGACTGTCGAGGAATATGTTCATGCCATGGAATTACTAGTGAATGATGCTCGTTTTACCCTTTACAACATATGTTATAAGAGAGTTCTTGTTTGTTGGATTAGCCTTGCATTTCTCGTGCTTTTGGCACTGCTGTTTTCAGGGTTAACAGGTTTGACCTTGTTTTCCCTAGGAGTACTGTGGCTCATTTTCAATGCTGCTGCTATATTTCTCTGTATGTGGATAAAATTAAAATTATCTAAAGGATTGGACCAATGCCTGGCAAGAGTAAACAAACTATTAAACAAACATAAATTGATTTTGGCCTTAGATGATAGGGGTAAAATTTCATGTCATAAAGTGAACTTGTGTTTCATATATTTTGACTCTGGTCCTTGTATTATGCACATACAACAATTCATTGACAGTGAAGAAGGAAAAACTATAATGCAAGGTTGGGAGCAACGACTAGATATTACTACTAATGACATAGTTATTCAAGGGAGCCAAACTACTAGGTTATCACGTAAACAGGAACGGGGCGCGCTTCTATTTTTACGTTACGCTGCAAGATGGGGCGCGTATGTTCTCAAAAACAGATTGAATCTCGGTACGAATGTACCTGGCCGACACGGACATAAGTATGTCTGTCCATGTCAATACATAGAAGATCACCTCAAAGCAAAGCCACCCACAGGTATAACCAAGTATTTCTCACTGTCTAATCAAAATAATGAAATGTATTAA
Protein
MNNADNSIEFSEVNERNGNVHEEKIRKGWVQFDDEQQTPTVSHSENEIAVTKTPPVASTQTPTRPTVPAVLNTETVHVNLERGDKNLESSTQSTLTKNVEFVNVRHGFSNGDIIVTLLPVNTKWPWITAAQFRPELVPEELMAQGLTLTVEEYVHAMELLVNDARFTLYNICYKRVLVCWISLAFLVLLALLFSGLTGLTLFSLGVLWLIFNAAAIFLCMWIKLKLSKGLDQCLARVNKLLNKHKLILALDDRGKISCHKVNLCFIYFDSGPCIMHIQQFIDSEEGKTIMQGWEQRLDITTNDIVIQGSQTTRLSRKQERGALLFLRYAARWGAYVLKNRLNLGTNVPGRHGHKYVCPCQYIEDHLKAKPPTGITKYFSLSNQNNEMY
Summary
Uniprot
H9J9K6
A0A2A4IXG8
A0A194PLL3
A0A2W1C011
A0A2H1WS40
A0A2A4IW03
+ More
A0A1E1VXN8 A0A194R1V7 A0A2A4IVT6 A0A0L7LH56 A0A1Y1L8Q6 A0A1Y1L678 Q16GK7 A0A0K8TKM3 A0A212F0P0 A0A1Q3FNC0 Q7Q1E1 A0A1Q3FN13 A0A182GKE1 B0WPF9 A0A2M4CR36 W5JDZ7 A0A2M4CQN1 A0A2M3ZFS8 A0A1W4XAE3 U4UYE3 A0A023ESN8 A0A1Y1L6I2 D6WKM5 N6T808 A0A1L8D9E8 A0A0R3NXA5 A0A1L8DJX8 A0A3B0JJ94 A0A0R3NWM4 A0A0P6IY54 A0A336MXB7 A0A336MYV8 A0A1W4V8N6 A0A0Q9W9Z3 A0A1W4V7S9 A0A1W4UV00 A0A0Q9VYY4 A0A1W4V7A8 M9PBB3 A0A0Q9WR31 M9PCX0 D6W4W6 M9PFU6 A0A1W4UVD4 A0A0J9R230 A0A182QXA8 Q9VJW7 A0A0Q5WNM1 A0A0J9R2V0 A0A182NC06 A0A0J9R200 A0A0Q5W9Z6 A0A0R1DK62 B4IEP7 A0A1B6CD06 A0A0J9TMW3 A0A0Q5WAG6 A0A026VW89 A0A1B6G905 A0A1B6GPT3 A0A1B6M225 A0A1B6DHH4 A0A0R1DSS5 A0A1B6KN43 A0A0Q5WJI6 A0A0P8ZHW9 A0A1B6D563 A0A0P8XHJ0 A0A195CDK9 A0A084W8T5 A0A182MMS5 A0A182JLJ1 A0A0P8XXW9 A0A2M4BN66 A0A182YDI8 A0A2M4A4J2 A0A2M4BN75 A0A1B0D8S7 A0A2M4BNG8 A0A336LC98 A0A1Q3G4U7 A0A1W7R7Y8 A0A2M4A581
A0A1E1VXN8 A0A194R1V7 A0A2A4IVT6 A0A0L7LH56 A0A1Y1L8Q6 A0A1Y1L678 Q16GK7 A0A0K8TKM3 A0A212F0P0 A0A1Q3FNC0 Q7Q1E1 A0A1Q3FN13 A0A182GKE1 B0WPF9 A0A2M4CR36 W5JDZ7 A0A2M4CQN1 A0A2M3ZFS8 A0A1W4XAE3 U4UYE3 A0A023ESN8 A0A1Y1L6I2 D6WKM5 N6T808 A0A1L8D9E8 A0A0R3NXA5 A0A1L8DJX8 A0A3B0JJ94 A0A0R3NWM4 A0A0P6IY54 A0A336MXB7 A0A336MYV8 A0A1W4V8N6 A0A0Q9W9Z3 A0A1W4V7S9 A0A1W4UV00 A0A0Q9VYY4 A0A1W4V7A8 M9PBB3 A0A0Q9WR31 M9PCX0 D6W4W6 M9PFU6 A0A1W4UVD4 A0A0J9R230 A0A182QXA8 Q9VJW7 A0A0Q5WNM1 A0A0J9R2V0 A0A182NC06 A0A0J9R200 A0A0Q5W9Z6 A0A0R1DK62 B4IEP7 A0A1B6CD06 A0A0J9TMW3 A0A0Q5WAG6 A0A026VW89 A0A1B6G905 A0A1B6GPT3 A0A1B6M225 A0A1B6DHH4 A0A0R1DSS5 A0A1B6KN43 A0A0Q5WJI6 A0A0P8ZHW9 A0A1B6D563 A0A0P8XHJ0 A0A195CDK9 A0A084W8T5 A0A182MMS5 A0A182JLJ1 A0A0P8XXW9 A0A2M4BN66 A0A182YDI8 A0A2M4A4J2 A0A2M4BN75 A0A1B0D8S7 A0A2M4BNG8 A0A336LC98 A0A1Q3G4U7 A0A1W7R7Y8 A0A2M4A581
Pubmed
19121390
26354079
28756777
26227816
28004739
17510324
+ More
26369729 22118469 12364791 14747013 17210077 26483478 20920257 23761445 23537049 24945155 18362917 19820115 15632085 17994087 26999592 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 24508170 24438588 25244985
26369729 22118469 12364791 14747013 17210077 26483478 20920257 23761445 23537049 24945155 18362917 19820115 15632085 17994087 26999592 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 17550304 24508170 24438588 25244985
EMBL
BABH01005001
NWSH01005932
PCG63840.1
KQ459601
KPI93888.1
KZ149895
+ More
PZC78847.1 ODYU01010624 SOQ55858.1 PCG63839.1 GDQN01011577 JAT79477.1 KQ460870 KPJ11768.1 PCG63841.1 JTDY01001163 KOB74694.1 GEZM01063235 JAV69171.1 GEZM01063234 JAV69172.1 CH478268 EAT33367.1 GDAI01002691 JAI14912.1 AGBW02011047 OWR47316.1 GFDL01005988 JAV29057.1 AAAB01008980 EAA14193.4 GFDL01006066 JAV28979.1 JXUM01069816 KQ562571 KXJ75578.1 DS232023 EDS32338.1 GGFL01003463 MBW67641.1 ADMH02001500 ETN62296.1 GGFL01003462 MBW67640.1 GGFM01006529 MBW27280.1 KB632427 ERL95391.1 GAPW01001310 JAC12288.1 GEZM01063233 GEZM01063232 JAV69174.1 KQ971342 EFA03018.1 APGK01040533 APGK01040534 APGK01040535 KB740982 ENN76369.1 GFDF01011003 JAV03081.1 CH379058 KRT03443.1 GFDF01007440 JAV06644.1 OUUW01000006 SPP82315.1 KRT03445.1 GDUN01000185 JAN95734.1 UFQT01003293 SSX34820.1 SSX34821.1 CH940654 KRF78036.1 KRF78035.1 AE014134 AGB92983.1 CH963920 KRF98692.1 AGB92984.1 BT125016 ADI46812.1 AGB92982.1 CM002910 KMY90138.1 AXCN02000699 AAF53321.2 CH954177 KQS70472.1 KMY90139.1 KMY90136.1 KQS70471.1 CM000157 KRJ97703.1 CH480831 EDW46074.1 GEDC01026148 JAS11150.1 KMY90140.1 KQS70470.1 KK107942 EZA47089.1 GECZ01010870 JAS58899.1 GECZ01005324 JAS64445.1 GEBQ01010033 JAT29944.1 GEDC01012165 JAS25133.1 KRJ97702.1 GEBQ01027115 JAT12862.1 KQS70473.1 CH902624 KPU74303.1 GEDC01016459 JAS20839.1 KPU74305.1 KQ978009 KYM98148.1 ATLV01021514 KE525319 KFB46629.1 AXCM01002731 KPU74304.1 GGFJ01005388 MBW54529.1 GGFK01002329 MBW35650.1 GGFJ01005389 MBW54530.1 AJVK01027701 GGFJ01005479 MBW54620.1 UFQS01003038 UFQT01003038 SSX15099.1 SSX34479.1 GFDL01000215 JAV34830.1 GEHC01000363 JAV47282.1 GGFK01002580 MBW35901.1
PZC78847.1 ODYU01010624 SOQ55858.1 PCG63839.1 GDQN01011577 JAT79477.1 KQ460870 KPJ11768.1 PCG63841.1 JTDY01001163 KOB74694.1 GEZM01063235 JAV69171.1 GEZM01063234 JAV69172.1 CH478268 EAT33367.1 GDAI01002691 JAI14912.1 AGBW02011047 OWR47316.1 GFDL01005988 JAV29057.1 AAAB01008980 EAA14193.4 GFDL01006066 JAV28979.1 JXUM01069816 KQ562571 KXJ75578.1 DS232023 EDS32338.1 GGFL01003463 MBW67641.1 ADMH02001500 ETN62296.1 GGFL01003462 MBW67640.1 GGFM01006529 MBW27280.1 KB632427 ERL95391.1 GAPW01001310 JAC12288.1 GEZM01063233 GEZM01063232 JAV69174.1 KQ971342 EFA03018.1 APGK01040533 APGK01040534 APGK01040535 KB740982 ENN76369.1 GFDF01011003 JAV03081.1 CH379058 KRT03443.1 GFDF01007440 JAV06644.1 OUUW01000006 SPP82315.1 KRT03445.1 GDUN01000185 JAN95734.1 UFQT01003293 SSX34820.1 SSX34821.1 CH940654 KRF78036.1 KRF78035.1 AE014134 AGB92983.1 CH963920 KRF98692.1 AGB92984.1 BT125016 ADI46812.1 AGB92982.1 CM002910 KMY90138.1 AXCN02000699 AAF53321.2 CH954177 KQS70472.1 KMY90139.1 KMY90136.1 KQS70471.1 CM000157 KRJ97703.1 CH480831 EDW46074.1 GEDC01026148 JAS11150.1 KMY90140.1 KQS70470.1 KK107942 EZA47089.1 GECZ01010870 JAS58899.1 GECZ01005324 JAS64445.1 GEBQ01010033 JAT29944.1 GEDC01012165 JAS25133.1 KRJ97702.1 GEBQ01027115 JAT12862.1 KQS70473.1 CH902624 KPU74303.1 GEDC01016459 JAS20839.1 KPU74305.1 KQ978009 KYM98148.1 ATLV01021514 KE525319 KFB46629.1 AXCM01002731 KPU74304.1 GGFJ01005388 MBW54529.1 GGFK01002329 MBW35650.1 GGFJ01005389 MBW54530.1 AJVK01027701 GGFJ01005479 MBW54620.1 UFQS01003038 UFQT01003038 SSX15099.1 SSX34479.1 GFDL01000215 JAV34830.1 GEHC01000363 JAV47282.1 GGFK01002580 MBW35901.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000008820
+ More
UP000007151 UP000007062 UP000069940 UP000249989 UP000002320 UP000000673 UP000192223 UP000030742 UP000007266 UP000019118 UP000001819 UP000268350 UP000192221 UP000008792 UP000000803 UP000007798 UP000075886 UP000008711 UP000075884 UP000002282 UP000001292 UP000053097 UP000007801 UP000078542 UP000030765 UP000075883 UP000075880 UP000076408 UP000092462
UP000007151 UP000007062 UP000069940 UP000249989 UP000002320 UP000000673 UP000192223 UP000030742 UP000007266 UP000019118 UP000001819 UP000268350 UP000192221 UP000008792 UP000000803 UP000007798 UP000075886 UP000008711 UP000075884 UP000002282 UP000001292 UP000053097 UP000007801 UP000078542 UP000030765 UP000075883 UP000075880 UP000076408 UP000092462
Pfam
PF14800 DUF4481
Interpro
IPR028054
DUF4481
ProteinModelPortal
H9J9K6
A0A2A4IXG8
A0A194PLL3
A0A2W1C011
A0A2H1WS40
A0A2A4IW03
+ More
A0A1E1VXN8 A0A194R1V7 A0A2A4IVT6 A0A0L7LH56 A0A1Y1L8Q6 A0A1Y1L678 Q16GK7 A0A0K8TKM3 A0A212F0P0 A0A1Q3FNC0 Q7Q1E1 A0A1Q3FN13 A0A182GKE1 B0WPF9 A0A2M4CR36 W5JDZ7 A0A2M4CQN1 A0A2M3ZFS8 A0A1W4XAE3 U4UYE3 A0A023ESN8 A0A1Y1L6I2 D6WKM5 N6T808 A0A1L8D9E8 A0A0R3NXA5 A0A1L8DJX8 A0A3B0JJ94 A0A0R3NWM4 A0A0P6IY54 A0A336MXB7 A0A336MYV8 A0A1W4V8N6 A0A0Q9W9Z3 A0A1W4V7S9 A0A1W4UV00 A0A0Q9VYY4 A0A1W4V7A8 M9PBB3 A0A0Q9WR31 M9PCX0 D6W4W6 M9PFU6 A0A1W4UVD4 A0A0J9R230 A0A182QXA8 Q9VJW7 A0A0Q5WNM1 A0A0J9R2V0 A0A182NC06 A0A0J9R200 A0A0Q5W9Z6 A0A0R1DK62 B4IEP7 A0A1B6CD06 A0A0J9TMW3 A0A0Q5WAG6 A0A026VW89 A0A1B6G905 A0A1B6GPT3 A0A1B6M225 A0A1B6DHH4 A0A0R1DSS5 A0A1B6KN43 A0A0Q5WJI6 A0A0P8ZHW9 A0A1B6D563 A0A0P8XHJ0 A0A195CDK9 A0A084W8T5 A0A182MMS5 A0A182JLJ1 A0A0P8XXW9 A0A2M4BN66 A0A182YDI8 A0A2M4A4J2 A0A2M4BN75 A0A1B0D8S7 A0A2M4BNG8 A0A336LC98 A0A1Q3G4U7 A0A1W7R7Y8 A0A2M4A581
A0A1E1VXN8 A0A194R1V7 A0A2A4IVT6 A0A0L7LH56 A0A1Y1L8Q6 A0A1Y1L678 Q16GK7 A0A0K8TKM3 A0A212F0P0 A0A1Q3FNC0 Q7Q1E1 A0A1Q3FN13 A0A182GKE1 B0WPF9 A0A2M4CR36 W5JDZ7 A0A2M4CQN1 A0A2M3ZFS8 A0A1W4XAE3 U4UYE3 A0A023ESN8 A0A1Y1L6I2 D6WKM5 N6T808 A0A1L8D9E8 A0A0R3NXA5 A0A1L8DJX8 A0A3B0JJ94 A0A0R3NWM4 A0A0P6IY54 A0A336MXB7 A0A336MYV8 A0A1W4V8N6 A0A0Q9W9Z3 A0A1W4V7S9 A0A1W4UV00 A0A0Q9VYY4 A0A1W4V7A8 M9PBB3 A0A0Q9WR31 M9PCX0 D6W4W6 M9PFU6 A0A1W4UVD4 A0A0J9R230 A0A182QXA8 Q9VJW7 A0A0Q5WNM1 A0A0J9R2V0 A0A182NC06 A0A0J9R200 A0A0Q5W9Z6 A0A0R1DK62 B4IEP7 A0A1B6CD06 A0A0J9TMW3 A0A0Q5WAG6 A0A026VW89 A0A1B6G905 A0A1B6GPT3 A0A1B6M225 A0A1B6DHH4 A0A0R1DSS5 A0A1B6KN43 A0A0Q5WJI6 A0A0P8ZHW9 A0A1B6D563 A0A0P8XHJ0 A0A195CDK9 A0A084W8T5 A0A182MMS5 A0A182JLJ1 A0A0P8XXW9 A0A2M4BN66 A0A182YDI8 A0A2M4A4J2 A0A2M4BN75 A0A1B0D8S7 A0A2M4BNG8 A0A336LC98 A0A1Q3G4U7 A0A1W7R7Y8 A0A2M4A581
Ontologies
PANTHER
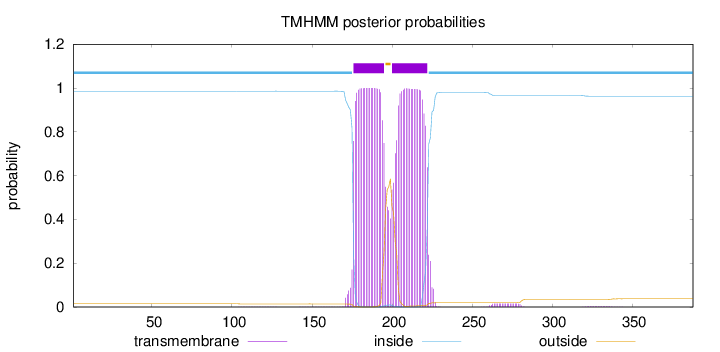
Topology
Length:
388
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
43.21424
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98561
inside
1 - 175
TMhelix
176 - 195
outside
196 - 199
TMhelix
200 - 222
inside
223 - 388
Population Genetic Test Statistics
Pi
196.87022
Theta
165.426614
Tajima's D
0.597803
CLR
42.741426
CSRT
0.548072596370181
Interpretation
Uncertain
