Gene
KWMTBOMO01886
Pre Gene Modal
BGIBMGA005947
Annotation
PREDICTED:_uncharacterized_protein_LOC101737309_isoform_X2_[Bombyx_mori]
Full name
Transcription termination factor, mitochondrial
Alternative Name
Mitochondrial transcription termination factor
Location in the cell
Nuclear Reliability : 3.075
Sequence
CDS
ATGAAAACATTGCTTCACATATACAAGGTCACTATGAATGATGTTGAAAATGGTTTCTGTGAAAACCGGCTTTATTATTTGGCATCTCGATTAAAGTGTTCACCAACTGGCTTATGTGAAAAATTAACTAAAAGAATCTTTGTTTACTCCTTATCATTTGATTGGTTGAAAAATGCCTTAGATATTTTGTTAGAAATGGGTGTGTCTGAAGACAGAATAATTCGGGATTTATGGATATTAAAATATCATCATGAAACAATACGCCTAAGACTTCAACAAGTCAAAGATAAGGGTATTGACAACTTATATCCTTGGATGGTTAGATGCCCTGAGGAGGTACTAAATAGGTCAATTAGATTAAGCCAAGAAACTAAAAATATTCTAGGAGAAACAAAGTCGACACAGATTTATTTAGCTAATCGTCTGAATACATCACTTGATTCTGTGGAAGAGCTGTACTCAAAAACACCTGCATTAAAAACAATCAGAGTGACCAAAGCAAAAAAGTTCCTAGATTTTCTTATTAATGAAGGATTTACTGTTGAAGACATCGCTAATAAATCAAGAGTGTTATCAGCTTCTCAAAAAACTGTACAGCAAAGATTGGAAATGTTACGTGATTTGGGTGTCAATGACATAAATTTGAATGTTCTTTGTAGAAGTAGAAGAGATTTTAAAAAGTATTGTGATACCTTACAATTACTTGCTAAAAAGGATAAATAG
Protein
MKTLLHIYKVTMNDVENGFCENRLYYLASRLKCSPTGLCEKLTKRIFVYSLSFDWLKNALDILLEMGVSEDRIIRDLWILKYHHETIRLRLQQVKDKGIDNLYPWMVRCPEEVLNRSIRLSQETKNILGETKSTQIYLANRLNTSLDSVEELYSKTPALKTIRVTKAKKFLDFLINEGFTVEDIANKSRVLSASQKTVQQRLEMLRDLGVNDINLNVLCRSRRDFKKYCDTLQLLAKKDK
Summary
Description
Transcription termination factor (PubMed:12626700, PubMed:15845400, PubMed:16648357). Binds promoter DNA and regulates mitochondrial replication and transcription (PubMed:12626700, PubMed:15845400, PubMed:16648357, PubMed:24068965). Transcription termination activity may be polarized with highest termination activity occurring when its DNA-binding site is positioned in the reverse orientation with respect to the incoming RNA polymerase (PubMed:15845400). Required for normal topology and maintenance of mitochondrial DNA (mtDNA) levels (PubMed:24068965). Regulates mtDNA replication by promoting replication pausing, possibly by acting as a natural barrier to replication fork progression (PubMed:24068965). Its function in replication pausing prevents unregulated replication that may occur for example by collisions between the machineries of DNA replication and transcription during mtDNA synthesis (PubMed:24068965). This ensures the incorporation of RNA transcripts into replication intermediates at the replication fork and allow for proper fork progression (PubMed:24068965). Shares mtDNA binding sites with the mitochondrial termination factor mTerf5 and thereby may antagonize mTerf5 function during replication to regulate pausing (PubMed:22784680, PubMed:24068965). Likely to function downstream of Dref which activates genes involved in mtDNA replication and maintenance (PubMed:19032147, PubMed:24068965).
Similarity
Belongs to the mTERF family.
Keywords
Complete proteome
DNA replication
Mitochondrion
Reference proteome
Transcription
Transcription regulation
Transit peptide
Feature
chain Transcription termination factor, mitochondrial
Uniprot
H9J8V5
A0A2H1W9A7
A0A2A4JBQ4
A0A2A4JB43
A0A1E1W7M0
A0A1E1WRW3
+ More
A0A1E1WMV2 A0A212F0R6 A0A1E1W075 I4DPM5 W8BAU0 A0A0J9R2C6 B4HXG1 Q9V3F3 B3MKK1 B4KEI8 A0A0A1XC43 B4MZB4 B4P0K1 A0A0K8URD3 A0A194PMD2 B4JEB0 A0A034VYE0 A0A3B0KBQ3 A0A1W4UW90 B3N9T1 A0A0M3QU30 B4M8S1 Q29KB5 B4GX69 A0A1B0DHV8 A0A1J1INZ3 B4Q5I6 A0A1L8E036 A0A0L0BZ47 A0A1B0CAM7 A0A067RC32 A0A1I8MZ29 A0A1A9VQC6 A0A1A9X634 A0A1B0BIX3 A0A182H6I7 A0A1B0FKW8 Q16TT0 A0A1J1IHY1 A0A1Q3F9B0 A0A1A9VZM1 B0WBZ9 A0A1I8QBK8 A0A2S2Q4L3 A0A182P686 Q7PWR6 A0A2J7QCP4 A0A182LMZ6 A0A182V726 A0A182I765 A0A182XDD7 A0A1S4H088 A0A182QGG9 A0A182TXE7 A0A182NGK7 A0A182F5R4 A0A182RUR2 A0A182M8S3 A0A182VU96 A0A182Y943 W5J488 A0A2S2PSK2 A0A182SIY5 J9K150 A0A182JSD7 A0A084VYM4 C4WVK7 A0A336LM22 A0A182JFU3 A0A1Y1MAU0 A0A1B6IH65 A0A1W4WDC3 A0A293LNZ5 A0A0K8SF31 A0A146M9E4 A0A2R5LLH0 A0A0T6BE24 A0A0L7LGN5 A0A1D2N964 A0A026WIZ3 A0A2P8Y7N6 A0A1S3D0R3 A0A1W4WPH9 A0A087TTM2 A0A195FB18
A0A1E1WMV2 A0A212F0R6 A0A1E1W075 I4DPM5 W8BAU0 A0A0J9R2C6 B4HXG1 Q9V3F3 B3MKK1 B4KEI8 A0A0A1XC43 B4MZB4 B4P0K1 A0A0K8URD3 A0A194PMD2 B4JEB0 A0A034VYE0 A0A3B0KBQ3 A0A1W4UW90 B3N9T1 A0A0M3QU30 B4M8S1 Q29KB5 B4GX69 A0A1B0DHV8 A0A1J1INZ3 B4Q5I6 A0A1L8E036 A0A0L0BZ47 A0A1B0CAM7 A0A067RC32 A0A1I8MZ29 A0A1A9VQC6 A0A1A9X634 A0A1B0BIX3 A0A182H6I7 A0A1B0FKW8 Q16TT0 A0A1J1IHY1 A0A1Q3F9B0 A0A1A9VZM1 B0WBZ9 A0A1I8QBK8 A0A2S2Q4L3 A0A182P686 Q7PWR6 A0A2J7QCP4 A0A182LMZ6 A0A182V726 A0A182I765 A0A182XDD7 A0A1S4H088 A0A182QGG9 A0A182TXE7 A0A182NGK7 A0A182F5R4 A0A182RUR2 A0A182M8S3 A0A182VU96 A0A182Y943 W5J488 A0A2S2PSK2 A0A182SIY5 J9K150 A0A182JSD7 A0A084VYM4 C4WVK7 A0A336LM22 A0A182JFU3 A0A1Y1MAU0 A0A1B6IH65 A0A1W4WDC3 A0A293LNZ5 A0A0K8SF31 A0A146M9E4 A0A2R5LLH0 A0A0T6BE24 A0A0L7LGN5 A0A1D2N964 A0A026WIZ3 A0A2P8Y7N6 A0A1S3D0R3 A0A1W4WPH9 A0A087TTM2 A0A195FB18
Pubmed
19121390
22118469
22651552
24495485
22936249
17994087
+ More
12626700 10731132 12537572 12537569 15845400 16648357 19032147 22784680 24068965 18057021 25830018 17550304 26354079 25348373 15632085 26108605 24845553 25315136 26483478 17510324 12364791 20966253 25244985 20920257 23761445 24438588 28004739 26823975 26227816 27289101 24508170 30249741 29403074
12626700 10731132 12537572 12537569 15845400 16648357 19032147 22784680 24068965 18057021 25830018 17550304 26354079 25348373 15632085 26108605 24845553 25315136 26483478 17510324 12364791 20966253 25244985 20920257 23761445 24438588 28004739 26823975 26227816 27289101 24508170 30249741 29403074
EMBL
BABH01004994
ODYU01007034
SOQ49422.1
NWSH01002168
PCG68984.1
PCG68986.1
+ More
GDQN01008203 JAT82851.1 GDQN01001304 JAT89750.1 GDQN01002739 JAT88315.1 AGBW02011047 OWR47322.1 GDQN01010712 JAT80342.1 AK403678 BAM19865.1 GAMC01008175 JAB98380.1 CM002910 KMY90238.1 CH480818 EDW51741.1 AY196479 AE014134 AY075225 AAL68092.1 AAO38861.1 CH902620 EDV31554.1 CH933807 EDW11867.1 KRG02957.1 GBXI01005772 JAD08520.1 CH963913 EDW77387.1 CM000157 EDW88995.1 GDHF01023163 JAI29151.1 KQ459601 KPI93894.1 CH916368 EDW03630.1 GAKP01010636 JAC48316.1 OUUW01000006 SPP82481.1 CH954177 EDV58576.1 CP012523 ALC39943.1 CH940654 EDW57597.1 CH379061 EAL33260.2 CH479195 EDW27396.1 AJVK01061659 CVRI01000055 CRL01276.1 CM000361 EDX05030.1 GFDF01001966 JAV12118.1 JRES01001125 KNC25308.1 AJWK01004116 KK852561 KDR21317.1 JXJN01015223 JXUM01114158 KQ565759 KXJ70692.1 CCAG010013161 CH477641 EAT37912.1 CVRI01000049 CRK99140.1 GFDL01010957 JAV24088.1 DS231884 EDS43059.1 GGMS01003451 MBY72654.1 AAAB01008984 EAA14996.4 NEVH01016288 PNF26345.1 APCN01003368 AXCN02002086 AXCM01000336 ADMH02002160 ETN58188.1 GGMR01019764 MBY32383.1 ABLF02032773 ATLV01018379 KE525231 KFB43068.1 AK341566 BAH71927.1 UFQT01000006 SSX17407.1 GEZM01038284 JAV81680.1 GECU01037121 GECU01021975 GECU01021450 GECU01018158 GECU01014831 JAS70585.1 JAS85731.1 JAS86256.1 JAS89548.1 JAS92875.1 GFWV01003715 MAA28445.1 GBRD01014112 JAG51714.1 GDHC01002957 JAQ15672.1 GGLE01006041 MBY10167.1 LJIG01001567 KRT85373.1 JTDY01001163 KOB74688.1 LJIJ01000138 ODN01803.1 KK107182 QOIP01000011 EZA55963.1 RLU17039.1 PYGN01000830 PSN40269.1 KK116685 KFM68461.1 KQ981693 KYN37623.1
GDQN01008203 JAT82851.1 GDQN01001304 JAT89750.1 GDQN01002739 JAT88315.1 AGBW02011047 OWR47322.1 GDQN01010712 JAT80342.1 AK403678 BAM19865.1 GAMC01008175 JAB98380.1 CM002910 KMY90238.1 CH480818 EDW51741.1 AY196479 AE014134 AY075225 AAL68092.1 AAO38861.1 CH902620 EDV31554.1 CH933807 EDW11867.1 KRG02957.1 GBXI01005772 JAD08520.1 CH963913 EDW77387.1 CM000157 EDW88995.1 GDHF01023163 JAI29151.1 KQ459601 KPI93894.1 CH916368 EDW03630.1 GAKP01010636 JAC48316.1 OUUW01000006 SPP82481.1 CH954177 EDV58576.1 CP012523 ALC39943.1 CH940654 EDW57597.1 CH379061 EAL33260.2 CH479195 EDW27396.1 AJVK01061659 CVRI01000055 CRL01276.1 CM000361 EDX05030.1 GFDF01001966 JAV12118.1 JRES01001125 KNC25308.1 AJWK01004116 KK852561 KDR21317.1 JXJN01015223 JXUM01114158 KQ565759 KXJ70692.1 CCAG010013161 CH477641 EAT37912.1 CVRI01000049 CRK99140.1 GFDL01010957 JAV24088.1 DS231884 EDS43059.1 GGMS01003451 MBY72654.1 AAAB01008984 EAA14996.4 NEVH01016288 PNF26345.1 APCN01003368 AXCN02002086 AXCM01000336 ADMH02002160 ETN58188.1 GGMR01019764 MBY32383.1 ABLF02032773 ATLV01018379 KE525231 KFB43068.1 AK341566 BAH71927.1 UFQT01000006 SSX17407.1 GEZM01038284 JAV81680.1 GECU01037121 GECU01021975 GECU01021450 GECU01018158 GECU01014831 JAS70585.1 JAS85731.1 JAS86256.1 JAS89548.1 JAS92875.1 GFWV01003715 MAA28445.1 GBRD01014112 JAG51714.1 GDHC01002957 JAQ15672.1 GGLE01006041 MBY10167.1 LJIG01001567 KRT85373.1 JTDY01001163 KOB74688.1 LJIJ01000138 ODN01803.1 KK107182 QOIP01000011 EZA55963.1 RLU17039.1 PYGN01000830 PSN40269.1 KK116685 KFM68461.1 KQ981693 KYN37623.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000001292
UP000000803
UP000007801
+ More
UP000009192 UP000007798 UP000002282 UP000053268 UP000001070 UP000268350 UP000192221 UP000008711 UP000092553 UP000008792 UP000001819 UP000008744 UP000092462 UP000183832 UP000000304 UP000037069 UP000092461 UP000027135 UP000095301 UP000078200 UP000092443 UP000092460 UP000069940 UP000249989 UP000092444 UP000008820 UP000091820 UP000002320 UP000095300 UP000075885 UP000007062 UP000235965 UP000075882 UP000075903 UP000075840 UP000076407 UP000075886 UP000075902 UP000075884 UP000069272 UP000075900 UP000075883 UP000075920 UP000076408 UP000000673 UP000075901 UP000007819 UP000075881 UP000030765 UP000075880 UP000192223 UP000037510 UP000094527 UP000053097 UP000279307 UP000245037 UP000079169 UP000054359 UP000078541
UP000009192 UP000007798 UP000002282 UP000053268 UP000001070 UP000268350 UP000192221 UP000008711 UP000092553 UP000008792 UP000001819 UP000008744 UP000092462 UP000183832 UP000000304 UP000037069 UP000092461 UP000027135 UP000095301 UP000078200 UP000092443 UP000092460 UP000069940 UP000249989 UP000092444 UP000008820 UP000091820 UP000002320 UP000095300 UP000075885 UP000007062 UP000235965 UP000075882 UP000075903 UP000075840 UP000076407 UP000075886 UP000075902 UP000075884 UP000069272 UP000075900 UP000075883 UP000075920 UP000076408 UP000000673 UP000075901 UP000007819 UP000075881 UP000030765 UP000075880 UP000192223 UP000037510 UP000094527 UP000053097 UP000279307 UP000245037 UP000079169 UP000054359 UP000078541
Interpro
SUPFAM
SSF56281
SSF56281
Gene 3D
ProteinModelPortal
H9J8V5
A0A2H1W9A7
A0A2A4JBQ4
A0A2A4JB43
A0A1E1W7M0
A0A1E1WRW3
+ More
A0A1E1WMV2 A0A212F0R6 A0A1E1W075 I4DPM5 W8BAU0 A0A0J9R2C6 B4HXG1 Q9V3F3 B3MKK1 B4KEI8 A0A0A1XC43 B4MZB4 B4P0K1 A0A0K8URD3 A0A194PMD2 B4JEB0 A0A034VYE0 A0A3B0KBQ3 A0A1W4UW90 B3N9T1 A0A0M3QU30 B4M8S1 Q29KB5 B4GX69 A0A1B0DHV8 A0A1J1INZ3 B4Q5I6 A0A1L8E036 A0A0L0BZ47 A0A1B0CAM7 A0A067RC32 A0A1I8MZ29 A0A1A9VQC6 A0A1A9X634 A0A1B0BIX3 A0A182H6I7 A0A1B0FKW8 Q16TT0 A0A1J1IHY1 A0A1Q3F9B0 A0A1A9VZM1 B0WBZ9 A0A1I8QBK8 A0A2S2Q4L3 A0A182P686 Q7PWR6 A0A2J7QCP4 A0A182LMZ6 A0A182V726 A0A182I765 A0A182XDD7 A0A1S4H088 A0A182QGG9 A0A182TXE7 A0A182NGK7 A0A182F5R4 A0A182RUR2 A0A182M8S3 A0A182VU96 A0A182Y943 W5J488 A0A2S2PSK2 A0A182SIY5 J9K150 A0A182JSD7 A0A084VYM4 C4WVK7 A0A336LM22 A0A182JFU3 A0A1Y1MAU0 A0A1B6IH65 A0A1W4WDC3 A0A293LNZ5 A0A0K8SF31 A0A146M9E4 A0A2R5LLH0 A0A0T6BE24 A0A0L7LGN5 A0A1D2N964 A0A026WIZ3 A0A2P8Y7N6 A0A1S3D0R3 A0A1W4WPH9 A0A087TTM2 A0A195FB18
A0A1E1WMV2 A0A212F0R6 A0A1E1W075 I4DPM5 W8BAU0 A0A0J9R2C6 B4HXG1 Q9V3F3 B3MKK1 B4KEI8 A0A0A1XC43 B4MZB4 B4P0K1 A0A0K8URD3 A0A194PMD2 B4JEB0 A0A034VYE0 A0A3B0KBQ3 A0A1W4UW90 B3N9T1 A0A0M3QU30 B4M8S1 Q29KB5 B4GX69 A0A1B0DHV8 A0A1J1INZ3 B4Q5I6 A0A1L8E036 A0A0L0BZ47 A0A1B0CAM7 A0A067RC32 A0A1I8MZ29 A0A1A9VQC6 A0A1A9X634 A0A1B0BIX3 A0A182H6I7 A0A1B0FKW8 Q16TT0 A0A1J1IHY1 A0A1Q3F9B0 A0A1A9VZM1 B0WBZ9 A0A1I8QBK8 A0A2S2Q4L3 A0A182P686 Q7PWR6 A0A2J7QCP4 A0A182LMZ6 A0A182V726 A0A182I765 A0A182XDD7 A0A1S4H088 A0A182QGG9 A0A182TXE7 A0A182NGK7 A0A182F5R4 A0A182RUR2 A0A182M8S3 A0A182VU96 A0A182Y943 W5J488 A0A2S2PSK2 A0A182SIY5 J9K150 A0A182JSD7 A0A084VYM4 C4WVK7 A0A336LM22 A0A182JFU3 A0A1Y1MAU0 A0A1B6IH65 A0A1W4WDC3 A0A293LNZ5 A0A0K8SF31 A0A146M9E4 A0A2R5LLH0 A0A0T6BE24 A0A0L7LGN5 A0A1D2N964 A0A026WIZ3 A0A2P8Y7N6 A0A1S3D0R3 A0A1W4WPH9 A0A087TTM2 A0A195FB18
Ontologies
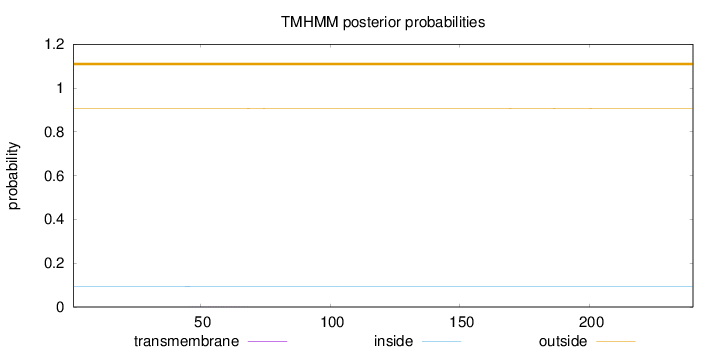
Topology
Subcellular location
Mitochondrion
Length:
240
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01
Exp number, first 60 AAs:
0.00682
Total prob of N-in:
0.09420
outside
1 - 240
Population Genetic Test Statistics
Pi
236.100501
Theta
187.433845
Tajima's D
0.812524
CLR
0.480275
CSRT
0.603469826508675
Interpretation
Possibly Positive selection
