Gene
KWMTBOMO01884 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005948
Annotation
PREDICTED:_C-type_lectin_4_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.315 Nuclear Reliability : 1.241 PlasmaMembrane Reliability : 1.032
Sequence
CDS
ATGAGCCGGCACTTTTGCCTGGCGGCAGCAGTTTTGTTCGCGGCACTATGCTTCGCTGAGGCCCAGAGACGTCTAGCTTTACCAGATCCCAGGAGTTGTGCCAACCGAGTACGCCATTCGACATACCGCGATGCACGGGGAGTACTACATTCGTATTTCTTTAGCTGGGAACATGCTCCAACACGCAGCTTAGAAGTGGATTGGCTGGACGCAAGGAACATCTGCCGCAGGCATTGCATGGACGCTGTTTCCTTAGAAACACCCCAGGAAAACGAATTTGTAAAGCAAAGGATCGCTCGCGGAAACATACGTTACATTTGGACTTCTGGTCGTAAATGTAATTTCGCCGGCTGTGACCGCGGTGACCTACAGCCTCCAAATGTGAACGGTTGGTTCTGGTCAGGTTCGGGAGCCAAGATCGGGCCCACCACCCAACGTAACACGGGCGATTGGTCTTACACCGGTGGCTACGGTCAAGCGCAGCCTGATAACAGAGAAGCTGCTCAAGGTAACGACGAGTCCTGTCTGGCGATACTTAACAACTTCTATAATGATGGCATTAAATGGCACGATGTGGCTTGCCACCACGTAAAGCCATTCGTGTGCGAAGACAGTGATGAACTACTTAATTTTGTCCGATCACGGAACCCGGGACTGCGTCTCTAA
Protein
MSRHFCLAAAVLFAALCFAEAQRRLALPDPRSCANRVRHSTYRDARGVLHSYFFSWEHAPTRSLEVDWLDARNICRRHCMDAVSLETPQENEFVKQRIARGNIRYIWTSGRKCNFAGCDRGDLQPPNVNGWFWSGSGAKIGPTTQRNTGDWSYTGGYGQAQPDNREAAQGNDESCLAILNNFYNDGIKWHDVACHHVKPFVCEDSDELLNFVRSRNPGLRL
Summary
Uniprot
D2X2F7
A0A2W1BK74
A0A2H1X2K3
A0A2A4JBB8
A0A212F0U5
A0A219YXG2
+ More
I4DLZ7 A0A2M4DNC3 A0A182M2E3 A0A182R6T5 A0A182YHA3 A0A182VT07 A0A182PF74 A0A182UWL9 A0A182JY93 A0A182TW34 A0A182XB44 A0A182KJQ4 Q5TPA6 A0A182I0N9 A0A182Q026 A0A182FMS9 A0A084W635 A0A194PKD1 T1DL69 A0A182N8K3 B2DBF4 A0A0L0CIK9 A0A1Q3FQW7 B0WF16 A0A1A9X8B0 A0A1B0BZL0 A0A1B0G6A1 A0A1B0AIT3 A0A1A9VA03 A0A023EIY2 A0A2K8JWW1 A0A1A9WBW0 A0A1I8Q8V5 A0A0A9ZAJ6 A0A1B0D1K8 A0A146MDY1 A0A1I8Q8W5 A0A182G3Y3 B3MKB4 A0A1I8NIN8 A0A0P8XGV6 A0A1J1I1Z7 W8C1K7 A0A0A1XEN8 A0A0K8TZL2 A0A034WC07 A0A1L8DQ56 B4KKA5 N6U964 B4LRV4 A0A2R7X319 A0A1W4W7D5 B4HY69 E0VF75 A0A0R1DLN5 B4Q5Q2 Q9VM07 A0A1B6EME2 A0A2A3E305 Q16R55 D3XL68 A0A087ZNY6 A0A3G1CIC9 B3N6K6 B4JDE4 D6WKP2 A0A067RBR3 A0A2P8Y7L9 A0A151X706 B4NZK1 E2B0H0 A0A2J7R7K2 B4MZU7 A0A1V1FIP4 F4W8F3 A0A195BAZ3 A0A158NK57 A0A195FAH5 A0A0M4EEX3 A0A1Y1L4E6 A0A195C2S6 A0A1S4H9W5 A0A195EIZ4 A0A3B0K7K5 Q29NX7 B4GJR1 A0A182T631 K7IQ55 A0A0M9A0H1 A0A171AVF2 T1H8X9 A0A1W4X6N4 A0A232ERG2 A0A0L7RAP9
I4DLZ7 A0A2M4DNC3 A0A182M2E3 A0A182R6T5 A0A182YHA3 A0A182VT07 A0A182PF74 A0A182UWL9 A0A182JY93 A0A182TW34 A0A182XB44 A0A182KJQ4 Q5TPA6 A0A182I0N9 A0A182Q026 A0A182FMS9 A0A084W635 A0A194PKD1 T1DL69 A0A182N8K3 B2DBF4 A0A0L0CIK9 A0A1Q3FQW7 B0WF16 A0A1A9X8B0 A0A1B0BZL0 A0A1B0G6A1 A0A1B0AIT3 A0A1A9VA03 A0A023EIY2 A0A2K8JWW1 A0A1A9WBW0 A0A1I8Q8V5 A0A0A9ZAJ6 A0A1B0D1K8 A0A146MDY1 A0A1I8Q8W5 A0A182G3Y3 B3MKB4 A0A1I8NIN8 A0A0P8XGV6 A0A1J1I1Z7 W8C1K7 A0A0A1XEN8 A0A0K8TZL2 A0A034WC07 A0A1L8DQ56 B4KKA5 N6U964 B4LRV4 A0A2R7X319 A0A1W4W7D5 B4HY69 E0VF75 A0A0R1DLN5 B4Q5Q2 Q9VM07 A0A1B6EME2 A0A2A3E305 Q16R55 D3XL68 A0A087ZNY6 A0A3G1CIC9 B3N6K6 B4JDE4 D6WKP2 A0A067RBR3 A0A2P8Y7L9 A0A151X706 B4NZK1 E2B0H0 A0A2J7R7K2 B4MZU7 A0A1V1FIP4 F4W8F3 A0A195BAZ3 A0A158NK57 A0A195FAH5 A0A0M4EEX3 A0A1Y1L4E6 A0A195C2S6 A0A1S4H9W5 A0A195EIZ4 A0A3B0K7K5 Q29NX7 B4GJR1 A0A182T631 K7IQ55 A0A0M9A0H1 A0A171AVF2 T1H8X9 A0A1W4X6N4 A0A232ERG2 A0A0L7RAP9
Pubmed
28756777
22118469
22651552
25244985
20966253
12364791
+ More
14747013 17210077 24438588 26354079 18712529 26108605 24945155 25401762 26823975 26483478 17994087 25315136 24495485 25830018 25348373 23537049 20566863 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 21695780 18362917 19820115 24845553 29403074 20798317 28410430 21719571 21347285 28004739 9087549 15632085 20075255 28648823
14747013 17210077 24438588 26354079 18712529 26108605 24945155 25401762 26823975 26483478 17994087 25315136 24495485 25830018 25348373 23537049 20566863 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 21695780 18362917 19820115 24845553 29403074 20798317 28410430 21719571 21347285 28004739 9087549 15632085 20075255 28648823
EMBL
GQ487490
ADB12588.1
KZ150033
PZC74691.1
ODYU01012907
SOQ59446.1
+ More
NWSH01002168 PCG68988.1 AGBW02011047 OWR47324.1 KX550116 AQY54433.1 AK402315 BAM18937.1 GGFL01014879 MBW79057.1 AXCM01000242 AAAB01008980 EAA13809.5 APCN01005226 AXCN02001838 ATLV01020721 KE525305 KFB45679.1 KQ459601 KPI93896.1 GAMD01000591 JAB01000.1 AB264633 BAG30713.1 JRES01000438 KNC31319.1 GFDL01005086 JAV29959.1 DS231913 EDS25924.1 JXJN01023188 JXJN01025268 CCAG010005175 GAPW01004185 JAC09413.1 KY031156 ATU82907.1 GBHO01002688 JAG40916.1 AJVK01010291 AJVK01010292 GDHC01000676 JAQ17953.1 JXUM01007001 KQ560228 KXJ83734.1 CH902620 EDV32498.2 KPU73979.1 CVRI01000038 CRK93786.1 GAMC01003512 GAMC01003511 JAC03044.1 GBXI01004443 JAD09849.1 GDHF01032606 GDHF01024141 GDHF01013501 JAI19708.1 JAI28173.1 JAI38813.1 GAKP01007096 GAKP01007095 JAC51857.1 GFDF01005488 JAV08596.1 CH933807 EDW12636.1 APGK01044125 KB741020 KB631628 ENN75122.1 ERL84836.1 CH940649 EDW64706.1 KK856583 PTY26051.1 CH480818 EDW51999.1 DS235107 EEB12031.1 CM000157 KRJ97373.1 CM000361 CM002910 EDX04098.1 KMY88780.1 AE014134 BT001650 AAF52521.1 AAN71405.1 GECZ01030695 JAS39074.1 KZ288443 PBC25692.1 CH477724 EAT36878.1 GU358193 ADD51171.1 KT808468 KT808469 ALG65085.1 CH954177 EDV59222.1 CH916368 EDW03314.1 KQ971342 EFA04534.1 KK852561 KDR21331.1 PYGN01000830 PSN40257.1 KQ982472 KYQ56078.1 EDW87750.2 GL444608 EFN60812.1 NEVH01006731 PNF36813.1 CH963920 EDW77882.1 FX985238 BAX07251.1 GL887908 EGI69444.1 KQ976537 KYM81390.1 ADTU01018706 KQ981693 KYN37625.1 CP012523 ALC38830.1 GEZM01066900 JAV67661.1 KQ978344 KYM94895.1 AAAB01007797 KQ978881 KYN27834.1 OUUW01000006 SPP81999.1 CH379058 EAL34516.1 CH479184 EDW36877.1 KQ435798 KOX73309.1 GEMB01000605 JAS02527.1 ACPB03001436 NNAY01002650 OXU20866.1 KQ414618 KOC67908.1
NWSH01002168 PCG68988.1 AGBW02011047 OWR47324.1 KX550116 AQY54433.1 AK402315 BAM18937.1 GGFL01014879 MBW79057.1 AXCM01000242 AAAB01008980 EAA13809.5 APCN01005226 AXCN02001838 ATLV01020721 KE525305 KFB45679.1 KQ459601 KPI93896.1 GAMD01000591 JAB01000.1 AB264633 BAG30713.1 JRES01000438 KNC31319.1 GFDL01005086 JAV29959.1 DS231913 EDS25924.1 JXJN01023188 JXJN01025268 CCAG010005175 GAPW01004185 JAC09413.1 KY031156 ATU82907.1 GBHO01002688 JAG40916.1 AJVK01010291 AJVK01010292 GDHC01000676 JAQ17953.1 JXUM01007001 KQ560228 KXJ83734.1 CH902620 EDV32498.2 KPU73979.1 CVRI01000038 CRK93786.1 GAMC01003512 GAMC01003511 JAC03044.1 GBXI01004443 JAD09849.1 GDHF01032606 GDHF01024141 GDHF01013501 JAI19708.1 JAI28173.1 JAI38813.1 GAKP01007096 GAKP01007095 JAC51857.1 GFDF01005488 JAV08596.1 CH933807 EDW12636.1 APGK01044125 KB741020 KB631628 ENN75122.1 ERL84836.1 CH940649 EDW64706.1 KK856583 PTY26051.1 CH480818 EDW51999.1 DS235107 EEB12031.1 CM000157 KRJ97373.1 CM000361 CM002910 EDX04098.1 KMY88780.1 AE014134 BT001650 AAF52521.1 AAN71405.1 GECZ01030695 JAS39074.1 KZ288443 PBC25692.1 CH477724 EAT36878.1 GU358193 ADD51171.1 KT808468 KT808469 ALG65085.1 CH954177 EDV59222.1 CH916368 EDW03314.1 KQ971342 EFA04534.1 KK852561 KDR21331.1 PYGN01000830 PSN40257.1 KQ982472 KYQ56078.1 EDW87750.2 GL444608 EFN60812.1 NEVH01006731 PNF36813.1 CH963920 EDW77882.1 FX985238 BAX07251.1 GL887908 EGI69444.1 KQ976537 KYM81390.1 ADTU01018706 KQ981693 KYN37625.1 CP012523 ALC38830.1 GEZM01066900 JAV67661.1 KQ978344 KYM94895.1 AAAB01007797 KQ978881 KYN27834.1 OUUW01000006 SPP81999.1 CH379058 EAL34516.1 CH479184 EDW36877.1 KQ435798 KOX73309.1 GEMB01000605 JAS02527.1 ACPB03001436 NNAY01002650 OXU20866.1 KQ414618 KOC67908.1
Proteomes
UP000218220
UP000007151
UP000075883
UP000075900
UP000076408
UP000075920
+ More
UP000075885 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075886 UP000069272 UP000030765 UP000053268 UP000075884 UP000037069 UP000002320 UP000092443 UP000092460 UP000092444 UP000092445 UP000078200 UP000091820 UP000095300 UP000092462 UP000069940 UP000249989 UP000007801 UP000095301 UP000183832 UP000009192 UP000019118 UP000030742 UP000008792 UP000192221 UP000001292 UP000009046 UP000002282 UP000000304 UP000000803 UP000242457 UP000008820 UP000005203 UP000008711 UP000001070 UP000007266 UP000027135 UP000245037 UP000075809 UP000000311 UP000235965 UP000007798 UP000007755 UP000078540 UP000005205 UP000078541 UP000092553 UP000078542 UP000078492 UP000268350 UP000001819 UP000008744 UP000075901 UP000002358 UP000053105 UP000015103 UP000192223 UP000215335 UP000053825
UP000075885 UP000075903 UP000075881 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000075886 UP000069272 UP000030765 UP000053268 UP000075884 UP000037069 UP000002320 UP000092443 UP000092460 UP000092444 UP000092445 UP000078200 UP000091820 UP000095300 UP000092462 UP000069940 UP000249989 UP000007801 UP000095301 UP000183832 UP000009192 UP000019118 UP000030742 UP000008792 UP000192221 UP000001292 UP000009046 UP000002282 UP000000304 UP000000803 UP000242457 UP000008820 UP000005203 UP000008711 UP000001070 UP000007266 UP000027135 UP000245037 UP000075809 UP000000311 UP000235965 UP000007798 UP000007755 UP000078540 UP000005205 UP000078541 UP000092553 UP000078542 UP000078492 UP000268350 UP000001819 UP000008744 UP000075901 UP000002358 UP000053105 UP000015103 UP000192223 UP000215335 UP000053825
Interpro
SUPFAM
SSF56436
SSF56436
Gene 3D
ProteinModelPortal
D2X2F7
A0A2W1BK74
A0A2H1X2K3
A0A2A4JBB8
A0A212F0U5
A0A219YXG2
+ More
I4DLZ7 A0A2M4DNC3 A0A182M2E3 A0A182R6T5 A0A182YHA3 A0A182VT07 A0A182PF74 A0A182UWL9 A0A182JY93 A0A182TW34 A0A182XB44 A0A182KJQ4 Q5TPA6 A0A182I0N9 A0A182Q026 A0A182FMS9 A0A084W635 A0A194PKD1 T1DL69 A0A182N8K3 B2DBF4 A0A0L0CIK9 A0A1Q3FQW7 B0WF16 A0A1A9X8B0 A0A1B0BZL0 A0A1B0G6A1 A0A1B0AIT3 A0A1A9VA03 A0A023EIY2 A0A2K8JWW1 A0A1A9WBW0 A0A1I8Q8V5 A0A0A9ZAJ6 A0A1B0D1K8 A0A146MDY1 A0A1I8Q8W5 A0A182G3Y3 B3MKB4 A0A1I8NIN8 A0A0P8XGV6 A0A1J1I1Z7 W8C1K7 A0A0A1XEN8 A0A0K8TZL2 A0A034WC07 A0A1L8DQ56 B4KKA5 N6U964 B4LRV4 A0A2R7X319 A0A1W4W7D5 B4HY69 E0VF75 A0A0R1DLN5 B4Q5Q2 Q9VM07 A0A1B6EME2 A0A2A3E305 Q16R55 D3XL68 A0A087ZNY6 A0A3G1CIC9 B3N6K6 B4JDE4 D6WKP2 A0A067RBR3 A0A2P8Y7L9 A0A151X706 B4NZK1 E2B0H0 A0A2J7R7K2 B4MZU7 A0A1V1FIP4 F4W8F3 A0A195BAZ3 A0A158NK57 A0A195FAH5 A0A0M4EEX3 A0A1Y1L4E6 A0A195C2S6 A0A1S4H9W5 A0A195EIZ4 A0A3B0K7K5 Q29NX7 B4GJR1 A0A182T631 K7IQ55 A0A0M9A0H1 A0A171AVF2 T1H8X9 A0A1W4X6N4 A0A232ERG2 A0A0L7RAP9
I4DLZ7 A0A2M4DNC3 A0A182M2E3 A0A182R6T5 A0A182YHA3 A0A182VT07 A0A182PF74 A0A182UWL9 A0A182JY93 A0A182TW34 A0A182XB44 A0A182KJQ4 Q5TPA6 A0A182I0N9 A0A182Q026 A0A182FMS9 A0A084W635 A0A194PKD1 T1DL69 A0A182N8K3 B2DBF4 A0A0L0CIK9 A0A1Q3FQW7 B0WF16 A0A1A9X8B0 A0A1B0BZL0 A0A1B0G6A1 A0A1B0AIT3 A0A1A9VA03 A0A023EIY2 A0A2K8JWW1 A0A1A9WBW0 A0A1I8Q8V5 A0A0A9ZAJ6 A0A1B0D1K8 A0A146MDY1 A0A1I8Q8W5 A0A182G3Y3 B3MKB4 A0A1I8NIN8 A0A0P8XGV6 A0A1J1I1Z7 W8C1K7 A0A0A1XEN8 A0A0K8TZL2 A0A034WC07 A0A1L8DQ56 B4KKA5 N6U964 B4LRV4 A0A2R7X319 A0A1W4W7D5 B4HY69 E0VF75 A0A0R1DLN5 B4Q5Q2 Q9VM07 A0A1B6EME2 A0A2A3E305 Q16R55 D3XL68 A0A087ZNY6 A0A3G1CIC9 B3N6K6 B4JDE4 D6WKP2 A0A067RBR3 A0A2P8Y7L9 A0A151X706 B4NZK1 E2B0H0 A0A2J7R7K2 B4MZU7 A0A1V1FIP4 F4W8F3 A0A195BAZ3 A0A158NK57 A0A195FAH5 A0A0M4EEX3 A0A1Y1L4E6 A0A195C2S6 A0A1S4H9W5 A0A195EIZ4 A0A3B0K7K5 Q29NX7 B4GJR1 A0A182T631 K7IQ55 A0A0M9A0H1 A0A171AVF2 T1H8X9 A0A1W4X6N4 A0A232ERG2 A0A0L7RAP9
PDB
5VC1
E-value=0.000639359,
Score=99
Ontologies
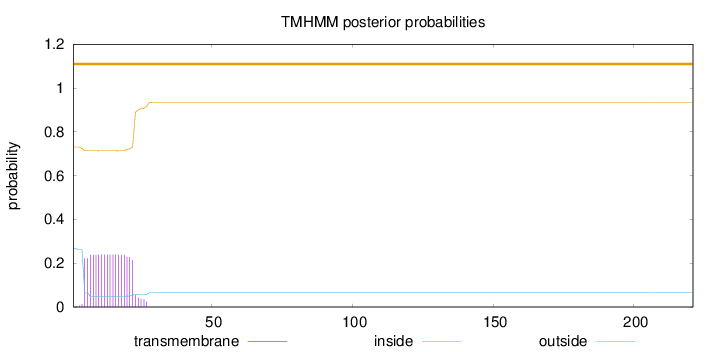
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.992602

Length:
221
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.43671999999999
Exp number, first 60 AAs:
4.43669
Total prob of N-in:
0.26960
outside
1 - 221
Population Genetic Test Statistics
Pi
180.229394
Theta
165.297271
Tajima's D
0.256792
CLR
0.829372
CSRT
0.43802809859507
Interpretation
Uncertain
