Gene
KWMTBOMO01880
Annotation
PREDICTED:_dolichyldiphosphatase_1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.571
Sequence
CDS
ATGTCGAAATTATTTGAAAATAAAATGGACAACGACCAAGTAAACGATTTCATGCATAAAAATGGCGATATCGAATGGCAGCCGTTAGCGCTTACTTTGGTTGAATATCCGAGAGGTGATTTAATTGGTAAATTCTTTGCATTCATCAGTTTATCTCCATTCGGTATTGGTGCTGGATTTGTAACCTTAATTTTGTTTCGTCGAGATCTTCACACTATAGCTTTCTTCTGTGGCACATTATTGAATGAAGCAGTTAATGCTACATTAAAGCATATTATTTGTGAATCTCGACCAATGACCAGAGGGAACTTGTATAATGAATATGGGATGCCTTCGTCACATGCTCAGTTTATTTGGTTTTTCAGTGTATATGTTTTTTATTTTGTTTTGATCAGATTACATCACATAAACAATAATAGCATAATATCGGCCTTATGGAGAGTTATAATTGTGAGTGGATGTATATGCCTAGCTGGACTTGTCAGCATTGGAAGAGTCTATTTGCATTACCACACCACTTCCCAAGTAGTGGTGGGGGCTATTGTAGGAATTATGTTTGCAACTATTTGGTTTGCACTGGTTCACAGAGTATTCACACCACTTTTCCCACAGATTGTTGCATTAAAAATAAGTGAAATGCTAATGATAAGGGACACAACGTTGATACCGAACGTATTGTGGTTCGAATACACGACGTCACGTCAAGAGGCTCGCGCTCGAGGCAGGAAGCTGGCGGCCCTGAAACCGACGCAATGA
Protein
MSKLFENKMDNDQVNDFMHKNGDIEWQPLALTLVEYPRGDLIGKFFAFISLSPFGIGAGFVTLILFRRDLHTIAFFCGTLLNEAVNATLKHIICESRPMTRGNLYNEYGMPSSHAQFIWFFSVYVFYFVLIRLHHINNNSIISALWRVIIVSGCICLAGLVSIGRVYLHYHTTSQVVVGAIVGIMFATIWFALVHRVFTPLFPQIVALKISEMLMIRDTTLIPNVLWFEYTTSRQEARARGRKLAALKPTQ
Summary
Uniprot
A0A2H1WD17
A0A2W1BP90
A0A1E1WIK2
A0A212F0P7
A0A194PKD6
A0A194R7A9
+ More
D6WLT1 A0A1Y1KDZ8 A0A0L7LA23 A0A2J7R703 A0A0T6BCW4 A0A2P8Y4S9 N6THK9 A0A1B6IPY5 A0A1B6LZU8 A0A1B6DK95 A0A1W4WNB2 A0A067RH86 A0A069DPT1 E0VVJ1 A0A0M9ABN5 A0A0K8TNR3 Q17E84 U5ES20 W5JS02 A0A2M4AJ17 A0A182FQ63 A0A2M3ZII7 A0A2M4BXD6 K7IMH3 A0A182MJY8 A0A182RQL7 A0A182VZY1 A0A182QC40 A0A182Y6R2 A0A182V2K6 A0A182P3W0 A0A182NR93 A0A1Q3F2V5 A0A1Q3F2U4 A0A154PJ09 A0A1Q3F2V9 A0A182HSB9 E2C341 A0A182XAI0 A0A182TPR0 Q7QCK1 A0A182K1G8 E2B250 A0A310SHX5 A0A0N7ZD59 A0A182LEA0 B0W990 A0A1B0CT93 A0A2S2N8K8 A0A0L7R8N1 A0A1L8DYC6 A0A195C7G0 J9JJ95 A0A2H8TXH5 A0A182IPW1 A0A146LGN3 A0A151IZ76 A0A195FL39 A0A3L8DNB3 A0A293LHF8 A0A087ZXL4 A0A2A3EC65 A0A1D2N0B9 T1IY84 E9IQ53 A0A1Z5L5S3 A0A1S3D840 A0A2R5LK76 A0A2T7NKE2 A0A1Z5L3B1 B7PT13 A0A0K8RMQ8 A0A131X9Q9 A0A224YW46 A0A131Z3T0 A0A023FRW4 A0A151I4Q5 A0A158NTZ2 A0A1E1X535 A0A023FZE8 U4U0S2 G3MPW7 L7MCR8 C3ZCJ1 A0A182TAQ2 A0A2M3ZBQ9 F7FFJ7 E9FSM1 M3YNG8 A0A2Y9J8I4 D2H158 G1MBT8 E2RQJ9 A0A2Y9H621
D6WLT1 A0A1Y1KDZ8 A0A0L7LA23 A0A2J7R703 A0A0T6BCW4 A0A2P8Y4S9 N6THK9 A0A1B6IPY5 A0A1B6LZU8 A0A1B6DK95 A0A1W4WNB2 A0A067RH86 A0A069DPT1 E0VVJ1 A0A0M9ABN5 A0A0K8TNR3 Q17E84 U5ES20 W5JS02 A0A2M4AJ17 A0A182FQ63 A0A2M3ZII7 A0A2M4BXD6 K7IMH3 A0A182MJY8 A0A182RQL7 A0A182VZY1 A0A182QC40 A0A182Y6R2 A0A182V2K6 A0A182P3W0 A0A182NR93 A0A1Q3F2V5 A0A1Q3F2U4 A0A154PJ09 A0A1Q3F2V9 A0A182HSB9 E2C341 A0A182XAI0 A0A182TPR0 Q7QCK1 A0A182K1G8 E2B250 A0A310SHX5 A0A0N7ZD59 A0A182LEA0 B0W990 A0A1B0CT93 A0A2S2N8K8 A0A0L7R8N1 A0A1L8DYC6 A0A195C7G0 J9JJ95 A0A2H8TXH5 A0A182IPW1 A0A146LGN3 A0A151IZ76 A0A195FL39 A0A3L8DNB3 A0A293LHF8 A0A087ZXL4 A0A2A3EC65 A0A1D2N0B9 T1IY84 E9IQ53 A0A1Z5L5S3 A0A1S3D840 A0A2R5LK76 A0A2T7NKE2 A0A1Z5L3B1 B7PT13 A0A0K8RMQ8 A0A131X9Q9 A0A224YW46 A0A131Z3T0 A0A023FRW4 A0A151I4Q5 A0A158NTZ2 A0A1E1X535 A0A023FZE8 U4U0S2 G3MPW7 L7MCR8 C3ZCJ1 A0A182TAQ2 A0A2M3ZBQ9 F7FFJ7 E9FSM1 M3YNG8 A0A2Y9J8I4 D2H158 G1MBT8 E2RQJ9 A0A2Y9H621
Pubmed
28756777
22118469
26354079
18362917
19820115
28004739
+ More
26227816 29403074 23537049 24845553 26334808 20566863 26369729 17510324 20920257 23761445 20075255 25244985 20798317 12364791 14747013 17210077 20966253 26823975 30249741 27289101 21282665 28528879 28049606 28797301 26830274 21347285 28503490 22216098 25576852 18563158 17495919 21292972 20010809 16341006
26227816 29403074 23537049 24845553 26334808 20566863 26369729 17510324 20920257 23761445 20075255 25244985 20798317 12364791 14747013 17210077 20966253 26823975 30249741 27289101 21282665 28528879 28049606 28797301 26830274 21347285 28503490 22216098 25576852 18563158 17495919 21292972 20010809 16341006
EMBL
ODYU01007836
SOQ50953.1
KZ150033
PZC74686.1
GDQN01004363
JAT86691.1
+ More
AGBW02011047 OWR47328.1 KQ459601 KPI93901.1 KQ460870 KPJ11756.1 KQ971343 EFA03406.1 GEZM01087849 JAV58440.1 JTDY01002006 KOB72348.1 NEVH01006738 PNF36615.1 LJIG01001772 KRT85168.1 PYGN01000933 PSN39241.1 APGK01037222 KB740948 KB632348 ENN77273.1 ERL93175.1 GECU01018759 JAS88947.1 GEBQ01010768 JAT29209.1 GEDC01011201 GEDC01011094 JAS26097.1 JAS26204.1 KK852470 KDR23196.1 GBGD01002811 JAC86078.1 DS235812 EEB17397.1 KQ435689 KOX81237.1 GDAI01002053 JAI15550.1 CH477284 EAT44794.1 GANO01002562 JAB57309.1 ADMH02000531 ETN66078.1 GGFK01007459 MBW40780.1 GGFM01007582 MBW28333.1 GGFJ01008596 MBW57737.1 AXCM01000211 AXCN02001467 GFDL01013150 JAV21895.1 GFDL01013160 JAV21885.1 KQ434931 KZC11839.1 GFDL01013149 JAV21896.1 APCN01000261 GL452274 EFN77657.1 AAAB01008859 EAA08176.3 GL445059 EFN60225.1 KQ763581 OAD54922.1 GDRN01052102 GDRN01052101 JAI66336.1 DS231862 EDS39787.1 AJWK01027296 GGMR01000487 MBY13106.1 KQ414631 KOC67230.1 GFDF01002675 JAV11409.1 KQ978143 KYM96782.1 ABLF02022047 GFXV01006063 MBW17868.1 GDHC01011305 JAQ07324.1 KQ980728 KYN14000.1 KQ981522 KYN40704.1 QOIP01000006 RLU21897.1 GFWV01002578 MAA27308.1 KZ288288 PBC29327.1 LJIJ01000327 ODM98747.1 JH431671 GL764659 EFZ17379.1 GFJQ02004203 JAW02767.1 GGLE01005798 MBY09924.1 PZQS01000011 PVD21629.1 GFJQ02005088 JAW01882.1 ABJB010472804 ABJB010773700 DS782609 EEC09735.1 GADI01001655 GEFM01003781 JAA72153.1 JAP72015.1 GEFH01005052 JAP63529.1 GFPF01007377 MAA18523.1 GEDV01002929 JAP85628.1 GBBK01001039 JAC23443.1 KQ976445 KYM86086.1 ADTU01026217 GFAC01004840 JAT94348.1 GBBL01000443 JAC26877.1 KB630512 ERL83645.1 JO843918 AEO35535.1 GACK01003249 JAA61785.1 GG666608 EEN49695.1 GGFM01005245 MBW25996.1 GL732524 EFX89802.1 AEYP01042563 AEYP01042564 GL192421 EFB25063.1 ACTA01121202 AAEX03006852
AGBW02011047 OWR47328.1 KQ459601 KPI93901.1 KQ460870 KPJ11756.1 KQ971343 EFA03406.1 GEZM01087849 JAV58440.1 JTDY01002006 KOB72348.1 NEVH01006738 PNF36615.1 LJIG01001772 KRT85168.1 PYGN01000933 PSN39241.1 APGK01037222 KB740948 KB632348 ENN77273.1 ERL93175.1 GECU01018759 JAS88947.1 GEBQ01010768 JAT29209.1 GEDC01011201 GEDC01011094 JAS26097.1 JAS26204.1 KK852470 KDR23196.1 GBGD01002811 JAC86078.1 DS235812 EEB17397.1 KQ435689 KOX81237.1 GDAI01002053 JAI15550.1 CH477284 EAT44794.1 GANO01002562 JAB57309.1 ADMH02000531 ETN66078.1 GGFK01007459 MBW40780.1 GGFM01007582 MBW28333.1 GGFJ01008596 MBW57737.1 AXCM01000211 AXCN02001467 GFDL01013150 JAV21895.1 GFDL01013160 JAV21885.1 KQ434931 KZC11839.1 GFDL01013149 JAV21896.1 APCN01000261 GL452274 EFN77657.1 AAAB01008859 EAA08176.3 GL445059 EFN60225.1 KQ763581 OAD54922.1 GDRN01052102 GDRN01052101 JAI66336.1 DS231862 EDS39787.1 AJWK01027296 GGMR01000487 MBY13106.1 KQ414631 KOC67230.1 GFDF01002675 JAV11409.1 KQ978143 KYM96782.1 ABLF02022047 GFXV01006063 MBW17868.1 GDHC01011305 JAQ07324.1 KQ980728 KYN14000.1 KQ981522 KYN40704.1 QOIP01000006 RLU21897.1 GFWV01002578 MAA27308.1 KZ288288 PBC29327.1 LJIJ01000327 ODM98747.1 JH431671 GL764659 EFZ17379.1 GFJQ02004203 JAW02767.1 GGLE01005798 MBY09924.1 PZQS01000011 PVD21629.1 GFJQ02005088 JAW01882.1 ABJB010472804 ABJB010773700 DS782609 EEC09735.1 GADI01001655 GEFM01003781 JAA72153.1 JAP72015.1 GEFH01005052 JAP63529.1 GFPF01007377 MAA18523.1 GEDV01002929 JAP85628.1 GBBK01001039 JAC23443.1 KQ976445 KYM86086.1 ADTU01026217 GFAC01004840 JAT94348.1 GBBL01000443 JAC26877.1 KB630512 ERL83645.1 JO843918 AEO35535.1 GACK01003249 JAA61785.1 GG666608 EEN49695.1 GGFM01005245 MBW25996.1 GL732524 EFX89802.1 AEYP01042563 AEYP01042564 GL192421 EFB25063.1 ACTA01121202 AAEX03006852
Proteomes
UP000007151
UP000053268
UP000053240
UP000007266
UP000037510
UP000235965
+ More
UP000245037 UP000019118 UP000030742 UP000192223 UP000027135 UP000009046 UP000053105 UP000008820 UP000000673 UP000069272 UP000002358 UP000075883 UP000075900 UP000075920 UP000075886 UP000076408 UP000075903 UP000075885 UP000075884 UP000076502 UP000075840 UP000008237 UP000076407 UP000075902 UP000007062 UP000075881 UP000000311 UP000075882 UP000002320 UP000092461 UP000053825 UP000078542 UP000007819 UP000075880 UP000078492 UP000078541 UP000279307 UP000005203 UP000242457 UP000094527 UP000079169 UP000245119 UP000001555 UP000078540 UP000005205 UP000001554 UP000075901 UP000002280 UP000000305 UP000000715 UP000248482 UP000008912 UP000002254 UP000248481
UP000245037 UP000019118 UP000030742 UP000192223 UP000027135 UP000009046 UP000053105 UP000008820 UP000000673 UP000069272 UP000002358 UP000075883 UP000075900 UP000075920 UP000075886 UP000076408 UP000075903 UP000075885 UP000075884 UP000076502 UP000075840 UP000008237 UP000076407 UP000075902 UP000007062 UP000075881 UP000000311 UP000075882 UP000002320 UP000092461 UP000053825 UP000078542 UP000007819 UP000075880 UP000078492 UP000078541 UP000279307 UP000005203 UP000242457 UP000094527 UP000079169 UP000245119 UP000001555 UP000078540 UP000005205 UP000001554 UP000075901 UP000002280 UP000000305 UP000000715 UP000248482 UP000008912 UP000002254 UP000248481
Pfam
PF01569 PAP2
Interpro
SUPFAM
SSF48317
SSF48317
ProteinModelPortal
A0A2H1WD17
A0A2W1BP90
A0A1E1WIK2
A0A212F0P7
A0A194PKD6
A0A194R7A9
+ More
D6WLT1 A0A1Y1KDZ8 A0A0L7LA23 A0A2J7R703 A0A0T6BCW4 A0A2P8Y4S9 N6THK9 A0A1B6IPY5 A0A1B6LZU8 A0A1B6DK95 A0A1W4WNB2 A0A067RH86 A0A069DPT1 E0VVJ1 A0A0M9ABN5 A0A0K8TNR3 Q17E84 U5ES20 W5JS02 A0A2M4AJ17 A0A182FQ63 A0A2M3ZII7 A0A2M4BXD6 K7IMH3 A0A182MJY8 A0A182RQL7 A0A182VZY1 A0A182QC40 A0A182Y6R2 A0A182V2K6 A0A182P3W0 A0A182NR93 A0A1Q3F2V5 A0A1Q3F2U4 A0A154PJ09 A0A1Q3F2V9 A0A182HSB9 E2C341 A0A182XAI0 A0A182TPR0 Q7QCK1 A0A182K1G8 E2B250 A0A310SHX5 A0A0N7ZD59 A0A182LEA0 B0W990 A0A1B0CT93 A0A2S2N8K8 A0A0L7R8N1 A0A1L8DYC6 A0A195C7G0 J9JJ95 A0A2H8TXH5 A0A182IPW1 A0A146LGN3 A0A151IZ76 A0A195FL39 A0A3L8DNB3 A0A293LHF8 A0A087ZXL4 A0A2A3EC65 A0A1D2N0B9 T1IY84 E9IQ53 A0A1Z5L5S3 A0A1S3D840 A0A2R5LK76 A0A2T7NKE2 A0A1Z5L3B1 B7PT13 A0A0K8RMQ8 A0A131X9Q9 A0A224YW46 A0A131Z3T0 A0A023FRW4 A0A151I4Q5 A0A158NTZ2 A0A1E1X535 A0A023FZE8 U4U0S2 G3MPW7 L7MCR8 C3ZCJ1 A0A182TAQ2 A0A2M3ZBQ9 F7FFJ7 E9FSM1 M3YNG8 A0A2Y9J8I4 D2H158 G1MBT8 E2RQJ9 A0A2Y9H621
D6WLT1 A0A1Y1KDZ8 A0A0L7LA23 A0A2J7R703 A0A0T6BCW4 A0A2P8Y4S9 N6THK9 A0A1B6IPY5 A0A1B6LZU8 A0A1B6DK95 A0A1W4WNB2 A0A067RH86 A0A069DPT1 E0VVJ1 A0A0M9ABN5 A0A0K8TNR3 Q17E84 U5ES20 W5JS02 A0A2M4AJ17 A0A182FQ63 A0A2M3ZII7 A0A2M4BXD6 K7IMH3 A0A182MJY8 A0A182RQL7 A0A182VZY1 A0A182QC40 A0A182Y6R2 A0A182V2K6 A0A182P3W0 A0A182NR93 A0A1Q3F2V5 A0A1Q3F2U4 A0A154PJ09 A0A1Q3F2V9 A0A182HSB9 E2C341 A0A182XAI0 A0A182TPR0 Q7QCK1 A0A182K1G8 E2B250 A0A310SHX5 A0A0N7ZD59 A0A182LEA0 B0W990 A0A1B0CT93 A0A2S2N8K8 A0A0L7R8N1 A0A1L8DYC6 A0A195C7G0 J9JJ95 A0A2H8TXH5 A0A182IPW1 A0A146LGN3 A0A151IZ76 A0A195FL39 A0A3L8DNB3 A0A293LHF8 A0A087ZXL4 A0A2A3EC65 A0A1D2N0B9 T1IY84 E9IQ53 A0A1Z5L5S3 A0A1S3D840 A0A2R5LK76 A0A2T7NKE2 A0A1Z5L3B1 B7PT13 A0A0K8RMQ8 A0A131X9Q9 A0A224YW46 A0A131Z3T0 A0A023FRW4 A0A151I4Q5 A0A158NTZ2 A0A1E1X535 A0A023FZE8 U4U0S2 G3MPW7 L7MCR8 C3ZCJ1 A0A182TAQ2 A0A2M3ZBQ9 F7FFJ7 E9FSM1 M3YNG8 A0A2Y9J8I4 D2H158 G1MBT8 E2RQJ9 A0A2Y9H621
Ontologies
PANTHER
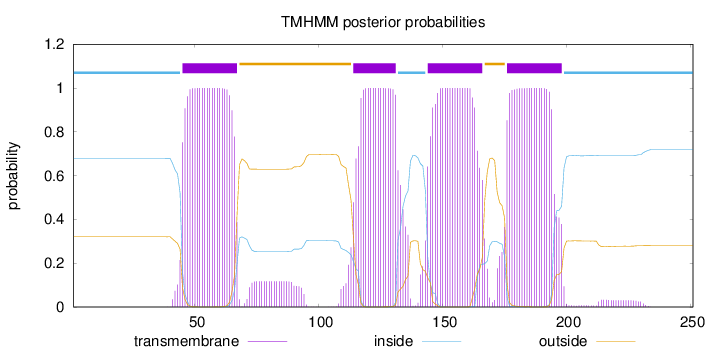
Topology
Length:
251
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.03399
Exp number, first 60 AAs:
16.03466
Total prob of N-in:
0.67915
POSSIBLE N-term signal
sequence
inside
1 - 44
TMhelix
45 - 67
outside
68 - 113
TMhelix
114 - 131
inside
132 - 143
TMhelix
144 - 166
outside
167 - 175
TMhelix
176 - 198
inside
199 - 251
Population Genetic Test Statistics
Pi
179.009474
Theta
152.068038
Tajima's D
0.188343
CLR
0.114919
CSRT
0.419229038548073
Interpretation
Uncertain
