Gene
KWMTBOMO01878 Validated by peptides from experiments
Annotation
coiled-coil_domain_containing_25_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.046 Nuclear Reliability : 2.438
Sequence
CDS
ATGGTGTTCTATTTTACAAGTAGTGTTGTATCTCCCCCAGTTACACTGTTTATGGGAGCCGACAAAAACGAAAATGAAGACTTAATAAAATGGGGTTGGCCAGAGGATGTCTGGTTTCATGTTGACAAAGTATCATCTGCACATGTATATTTAAGACTTACTCCTGGTCAAACGATTGATGATATACCTAATTTAGTTCTTGATGATGCTTGCCAGCTGGTCAAAGCAAATTCCATTTTAGGCAATAAAATGAATGACATAGACATAGTTTATACAATGTGGTCAAATTTGAAGAAGACTGCTGGAATGGAGGTTGGACAAGTAGCATTCCATAAAGACAGAGAAGTAAGAAAGGCGAAAGTGGCCAAAAGAAATAATGAAATAGTAAATAGGCTGAATAAAACAAAAACAGAATCATTTCCAGACCTCAGACAAGAAAGAGAGAATCGAGATAGATTAGAGAGAGAAGATAAAAAGAAAGTTTTAAGAGAAAAAAAGGAGAAAGAGAAAGAAGAAGAAAAACGAAAAAAGGAAGAGGCAGAACTCAGAAGCTACTCTACATTAATGAAAGCGGAAAATATGTCCACTAACTATGATGGTAATGATTCCGATGATTTTATGTGA
Protein
MVFYFTSSVVSPPVTLFMGADKNENEDLIKWGWPEDVWFHVDKVSSAHVYLRLTPGQTIDDIPNLVLDDACQLVKANSILGNKMNDIDIVYTMWSNLKKTAGMEVGQVAFHKDREVRKAKVAKRNNEIVNRLNKTKTESFPDLRQERENRDRLEREDKKKVLREKKEKEKEEEKRKKEEAELRSYSTLMKAENMSTNYDGNDSDDFM
Summary
Similarity
Belongs to the peptidase C2 family.
Uniprot
Q2F6C5
A0A2A4JG36
A0A2W1BSH6
A0A2H1WDB8
A0A194R230
A0A194PRZ2
+ More
A0A212F0V4 S4P9Q4 A0A0L7LIC1 A0A0P4VN21 R4FN45 A0A1B0CNH1 A0A1B6GQW5 A0A023EIG3 A0A023EJK7 A0A224XPC9 A0A1B6JH59 A0A182QU94 T1DNU5 A0A182TR81 W5J5V5 A0A1W4WXB4 A0A1B6LGF9 A0A2M3ZAH8 A0A2M4BZM8 Q16RD5 A0A182WVN4 A0A182I9B3 A0A182UX66 A0A182KXM5 Q7PQG6 A0A182JC92 B0X9K9 A0A084VW18 A0A182VXJ3 A0A1L8DAG1 A0A0K8TNB0 D6WKP3 A0A182T0G5 A0A336KM95 A0A182F804 A0A2M4APA0 A0A182KDZ5 U5EWE4 A0A182LT59 A0A182YIA8 A0A1Q3F3Y3 R4WT77 A0A182REV9 A0A1B6DHV3 A0A1Y1LW75 A0A146LH06 A0A0A9WRJ3 J3JV33 A0A182P903 A0A2J7QPN8 A0A0L0CIQ3 A0A1B0DKS3 A0A067RHM0 A0A131XLD7 E2ALT8 A0A023FJB1 A0A023FV84 A0A1E1XAP0 A0A224Y5B2 A0A131Z117 A0A023GFZ9 G3MML0 A0A1W7RAS3 A0A2P8Z968 A0A1Z5KYC7 F4W4F2 E2BAA5 A0A195EZ57 E0VL05 E9JC10 B7QCK4 A0A0K8RKU3 A0A195CD31 A0A293MU14 L7LU44 A0A2R5LDV2 A0A195DP35 A0A026W2B0 D3PHY0 A0A210PI60 A0A0P4W8E9 A0A151WZB8 C1BUX8 R7VC91 A0A3B0K5M5 A0A0K8VQH7 C1C271 B4N275 A0A034UZ34 A0A0C9RCF8 A0A1W4UVF8
A0A212F0V4 S4P9Q4 A0A0L7LIC1 A0A0P4VN21 R4FN45 A0A1B0CNH1 A0A1B6GQW5 A0A023EIG3 A0A023EJK7 A0A224XPC9 A0A1B6JH59 A0A182QU94 T1DNU5 A0A182TR81 W5J5V5 A0A1W4WXB4 A0A1B6LGF9 A0A2M3ZAH8 A0A2M4BZM8 Q16RD5 A0A182WVN4 A0A182I9B3 A0A182UX66 A0A182KXM5 Q7PQG6 A0A182JC92 B0X9K9 A0A084VW18 A0A182VXJ3 A0A1L8DAG1 A0A0K8TNB0 D6WKP3 A0A182T0G5 A0A336KM95 A0A182F804 A0A2M4APA0 A0A182KDZ5 U5EWE4 A0A182LT59 A0A182YIA8 A0A1Q3F3Y3 R4WT77 A0A182REV9 A0A1B6DHV3 A0A1Y1LW75 A0A146LH06 A0A0A9WRJ3 J3JV33 A0A182P903 A0A2J7QPN8 A0A0L0CIQ3 A0A1B0DKS3 A0A067RHM0 A0A131XLD7 E2ALT8 A0A023FJB1 A0A023FV84 A0A1E1XAP0 A0A224Y5B2 A0A131Z117 A0A023GFZ9 G3MML0 A0A1W7RAS3 A0A2P8Z968 A0A1Z5KYC7 F4W4F2 E2BAA5 A0A195EZ57 E0VL05 E9JC10 B7QCK4 A0A0K8RKU3 A0A195CD31 A0A293MU14 L7LU44 A0A2R5LDV2 A0A195DP35 A0A026W2B0 D3PHY0 A0A210PI60 A0A0P4W8E9 A0A151WZB8 C1BUX8 R7VC91 A0A3B0K5M5 A0A0K8VQH7 C1C271 B4N275 A0A034UZ34 A0A0C9RCF8 A0A1W4UVF8
Pubmed
28756777
26354079
22118469
23622113
26227816
27129103
+ More
24945155 20920257 23761445 17510324 20966253 12364791 14747013 17210077 24438588 26369729 18362917 19820115 25244985 23691247 28004739 26823975 25401762 22516182 23537049 26108605 24845553 28049606 20798317 28503490 28797301 26830274 22216098 29403074 28528879 21719571 20566863 21282665 25576852 24508170 28812685 23254933 17994087 25348373
24945155 20920257 23761445 17510324 20966253 12364791 14747013 17210077 24438588 26369729 18362917 19820115 25244985 23691247 28004739 26823975 25401762 22516182 23537049 26108605 24845553 28049606 20798317 28503490 28797301 26830274 22216098 29403074 28528879 21719571 20566863 21282665 25576852 24508170 28812685 23254933 17994087 25348373
EMBL
DQ311147
ABD36092.1
NWSH01001584
PCG70789.1
KZ150033
PZC74683.1
+ More
ODYU01007836 SOQ50956.1 KQ460870 KPJ11752.1 KQ459601 KPI93905.1 AGBW02011047 OWR47331.1 GAIX01003654 JAA88906.1 JTDY01001018 KOB75114.1 GDKW01002322 JAI54273.1 GAHY01000708 JAA76802.1 AJWK01020303 GECZ01004943 JAS64826.1 GAPW01004395 JAC09203.1 GAPW01004396 JAC09202.1 GFTR01003492 JAW12934.1 GECU01014278 GECU01009294 JAS93428.1 JAS98412.1 AXCN02000282 GAMD01002570 JAA99020.1 ADMH02002036 ETN59827.1 GEBQ01017259 JAT22718.1 GGFM01004761 MBW25512.1 GGFJ01009385 MBW58526.1 CH477716 CH477673 EAT36970.1 EAT37432.1 APCN01003250 AAAB01008888 EAA08860.5 DS232541 EDS43178.1 ATLV01017375 KE525164 KFB42162.1 GFDF01010631 JAV03453.1 GDAI01001726 JAI15877.1 KQ971342 EFA03024.1 UFQS01000259 UFQT01000259 SSX02160.1 SSX22537.1 GGFK01009241 MBW42562.1 GANO01000617 JAB59254.1 AXCM01003824 GFDL01012769 JAV22276.1 AK417932 BAN21147.1 GEDC01019908 GEDC01012032 JAS17390.1 JAS25266.1 GEZM01048418 GEZM01048417 JAV76580.1 GDHC01012402 JAQ06227.1 GBHO01034521 GBRD01016253 JAG09083.1 JAG49573.1 APGK01044117 BT127099 KB741020 KB631628 AEE62061.1 ENN75120.1 ERL84835.1 NEVH01012088 PNF30523.1 JRES01000340 KNC32130.1 AJVK01068349 KK852678 KDR18662.1 GEFH01000618 JAP67963.1 GL440629 EFN65608.1 GBBK01003734 JAC20748.1 GBBL01001926 JAC25394.1 GFAC01002870 JAT96318.1 GFPF01000759 MAA11905.1 GEDV01003433 JAP85124.1 GBBM01002571 JAC32847.1 JO843111 AEO34728.1 GFAH01000138 JAV48251.1 PYGN01000141 PSN53046.1 GFJQ02006915 JAW00055.1 GL887532 EGI70847.1 GL446696 EFN87360.1 KQ981905 KYN33590.1 DS235263 EEB14061.1 GL771642 EFZ09634.1 ABJB010197797 ABJB010859757 DS908087 EEC16576.1 GADI01002096 GEFM01005425 JAA71712.1 JAP70371.1 KQ977935 KYM98712.1 GFWV01019268 MAA43996.1 GACK01010660 JAA54374.1 GGLE01003523 MBY07649.1 KQ980713 KYN14249.1 KK107483 EZA50133.1 BT121236 HACA01018088 ADD38166.1 CDW35449.1 NEDP02076665 OWF36175.1 GDRN01064932 GDRN01064931 JAI64761.1 KQ982649 KYQ53107.1 BT078407 ACO12831.1 AMQN01019122 KB295338 ELU13300.1 OUUW01000021 SPP89504.1 GDHF01011211 JAI41103.1 BT080950 ACO15374.1 CH963925 EDW78464.1 GAKP01022731 JAC36221.1 GBYB01010762 JAG80529.1
ODYU01007836 SOQ50956.1 KQ460870 KPJ11752.1 KQ459601 KPI93905.1 AGBW02011047 OWR47331.1 GAIX01003654 JAA88906.1 JTDY01001018 KOB75114.1 GDKW01002322 JAI54273.1 GAHY01000708 JAA76802.1 AJWK01020303 GECZ01004943 JAS64826.1 GAPW01004395 JAC09203.1 GAPW01004396 JAC09202.1 GFTR01003492 JAW12934.1 GECU01014278 GECU01009294 JAS93428.1 JAS98412.1 AXCN02000282 GAMD01002570 JAA99020.1 ADMH02002036 ETN59827.1 GEBQ01017259 JAT22718.1 GGFM01004761 MBW25512.1 GGFJ01009385 MBW58526.1 CH477716 CH477673 EAT36970.1 EAT37432.1 APCN01003250 AAAB01008888 EAA08860.5 DS232541 EDS43178.1 ATLV01017375 KE525164 KFB42162.1 GFDF01010631 JAV03453.1 GDAI01001726 JAI15877.1 KQ971342 EFA03024.1 UFQS01000259 UFQT01000259 SSX02160.1 SSX22537.1 GGFK01009241 MBW42562.1 GANO01000617 JAB59254.1 AXCM01003824 GFDL01012769 JAV22276.1 AK417932 BAN21147.1 GEDC01019908 GEDC01012032 JAS17390.1 JAS25266.1 GEZM01048418 GEZM01048417 JAV76580.1 GDHC01012402 JAQ06227.1 GBHO01034521 GBRD01016253 JAG09083.1 JAG49573.1 APGK01044117 BT127099 KB741020 KB631628 AEE62061.1 ENN75120.1 ERL84835.1 NEVH01012088 PNF30523.1 JRES01000340 KNC32130.1 AJVK01068349 KK852678 KDR18662.1 GEFH01000618 JAP67963.1 GL440629 EFN65608.1 GBBK01003734 JAC20748.1 GBBL01001926 JAC25394.1 GFAC01002870 JAT96318.1 GFPF01000759 MAA11905.1 GEDV01003433 JAP85124.1 GBBM01002571 JAC32847.1 JO843111 AEO34728.1 GFAH01000138 JAV48251.1 PYGN01000141 PSN53046.1 GFJQ02006915 JAW00055.1 GL887532 EGI70847.1 GL446696 EFN87360.1 KQ981905 KYN33590.1 DS235263 EEB14061.1 GL771642 EFZ09634.1 ABJB010197797 ABJB010859757 DS908087 EEC16576.1 GADI01002096 GEFM01005425 JAA71712.1 JAP70371.1 KQ977935 KYM98712.1 GFWV01019268 MAA43996.1 GACK01010660 JAA54374.1 GGLE01003523 MBY07649.1 KQ980713 KYN14249.1 KK107483 EZA50133.1 BT121236 HACA01018088 ADD38166.1 CDW35449.1 NEDP02076665 OWF36175.1 GDRN01064932 GDRN01064931 JAI64761.1 KQ982649 KYQ53107.1 BT078407 ACO12831.1 AMQN01019122 KB295338 ELU13300.1 OUUW01000021 SPP89504.1 GDHF01011211 JAI41103.1 BT080950 ACO15374.1 CH963925 EDW78464.1 GAKP01022731 JAC36221.1 GBYB01010762 JAG80529.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
UP000092461
+ More
UP000075886 UP000075902 UP000000673 UP000192223 UP000008820 UP000076407 UP000075840 UP000075903 UP000075882 UP000007062 UP000075880 UP000002320 UP000030765 UP000075920 UP000007266 UP000075901 UP000069272 UP000075881 UP000075883 UP000076408 UP000075900 UP000019118 UP000030742 UP000075885 UP000235965 UP000037069 UP000092462 UP000027135 UP000000311 UP000245037 UP000007755 UP000008237 UP000078541 UP000009046 UP000001555 UP000078542 UP000078492 UP000053097 UP000242188 UP000075809 UP000014760 UP000268350 UP000007798 UP000192221
UP000075886 UP000075902 UP000000673 UP000192223 UP000008820 UP000076407 UP000075840 UP000075903 UP000075882 UP000007062 UP000075880 UP000002320 UP000030765 UP000075920 UP000007266 UP000075901 UP000069272 UP000075881 UP000075883 UP000076408 UP000075900 UP000019118 UP000030742 UP000075885 UP000235965 UP000037069 UP000092462 UP000027135 UP000000311 UP000245037 UP000007755 UP000008237 UP000078541 UP000009046 UP000001555 UP000078542 UP000078492 UP000053097 UP000242188 UP000075809 UP000014760 UP000268350 UP000007798 UP000192221
Interpro
ProteinModelPortal
Q2F6C5
A0A2A4JG36
A0A2W1BSH6
A0A2H1WDB8
A0A194R230
A0A194PRZ2
+ More
A0A212F0V4 S4P9Q4 A0A0L7LIC1 A0A0P4VN21 R4FN45 A0A1B0CNH1 A0A1B6GQW5 A0A023EIG3 A0A023EJK7 A0A224XPC9 A0A1B6JH59 A0A182QU94 T1DNU5 A0A182TR81 W5J5V5 A0A1W4WXB4 A0A1B6LGF9 A0A2M3ZAH8 A0A2M4BZM8 Q16RD5 A0A182WVN4 A0A182I9B3 A0A182UX66 A0A182KXM5 Q7PQG6 A0A182JC92 B0X9K9 A0A084VW18 A0A182VXJ3 A0A1L8DAG1 A0A0K8TNB0 D6WKP3 A0A182T0G5 A0A336KM95 A0A182F804 A0A2M4APA0 A0A182KDZ5 U5EWE4 A0A182LT59 A0A182YIA8 A0A1Q3F3Y3 R4WT77 A0A182REV9 A0A1B6DHV3 A0A1Y1LW75 A0A146LH06 A0A0A9WRJ3 J3JV33 A0A182P903 A0A2J7QPN8 A0A0L0CIQ3 A0A1B0DKS3 A0A067RHM0 A0A131XLD7 E2ALT8 A0A023FJB1 A0A023FV84 A0A1E1XAP0 A0A224Y5B2 A0A131Z117 A0A023GFZ9 G3MML0 A0A1W7RAS3 A0A2P8Z968 A0A1Z5KYC7 F4W4F2 E2BAA5 A0A195EZ57 E0VL05 E9JC10 B7QCK4 A0A0K8RKU3 A0A195CD31 A0A293MU14 L7LU44 A0A2R5LDV2 A0A195DP35 A0A026W2B0 D3PHY0 A0A210PI60 A0A0P4W8E9 A0A151WZB8 C1BUX8 R7VC91 A0A3B0K5M5 A0A0K8VQH7 C1C271 B4N275 A0A034UZ34 A0A0C9RCF8 A0A1W4UVF8
A0A212F0V4 S4P9Q4 A0A0L7LIC1 A0A0P4VN21 R4FN45 A0A1B0CNH1 A0A1B6GQW5 A0A023EIG3 A0A023EJK7 A0A224XPC9 A0A1B6JH59 A0A182QU94 T1DNU5 A0A182TR81 W5J5V5 A0A1W4WXB4 A0A1B6LGF9 A0A2M3ZAH8 A0A2M4BZM8 Q16RD5 A0A182WVN4 A0A182I9B3 A0A182UX66 A0A182KXM5 Q7PQG6 A0A182JC92 B0X9K9 A0A084VW18 A0A182VXJ3 A0A1L8DAG1 A0A0K8TNB0 D6WKP3 A0A182T0G5 A0A336KM95 A0A182F804 A0A2M4APA0 A0A182KDZ5 U5EWE4 A0A182LT59 A0A182YIA8 A0A1Q3F3Y3 R4WT77 A0A182REV9 A0A1B6DHV3 A0A1Y1LW75 A0A146LH06 A0A0A9WRJ3 J3JV33 A0A182P903 A0A2J7QPN8 A0A0L0CIQ3 A0A1B0DKS3 A0A067RHM0 A0A131XLD7 E2ALT8 A0A023FJB1 A0A023FV84 A0A1E1XAP0 A0A224Y5B2 A0A131Z117 A0A023GFZ9 G3MML0 A0A1W7RAS3 A0A2P8Z968 A0A1Z5KYC7 F4W4F2 E2BAA5 A0A195EZ57 E0VL05 E9JC10 B7QCK4 A0A0K8RKU3 A0A195CD31 A0A293MU14 L7LU44 A0A2R5LDV2 A0A195DP35 A0A026W2B0 D3PHY0 A0A210PI60 A0A0P4W8E9 A0A151WZB8 C1BUX8 R7VC91 A0A3B0K5M5 A0A0K8VQH7 C1C271 B4N275 A0A034UZ34 A0A0C9RCF8 A0A1W4UVF8
Ontologies
GO
PANTHER
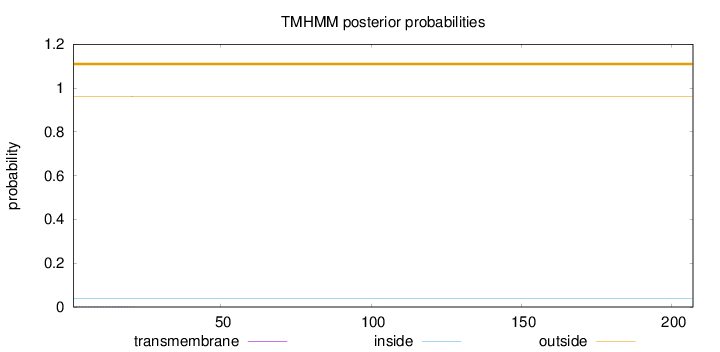
Topology
Length:
207
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03442
Exp number, first 60 AAs:
0.03397
Total prob of N-in:
0.03887
outside
1 - 207
Population Genetic Test Statistics
Pi
309.71329
Theta
218.768042
Tajima's D
1.316535
CLR
0
CSRT
0.746812659367032
Interpretation
Uncertain
