Gene
KWMTBOMO01877
Annotation
PREDICTED:_probable_queuine_tRNA-ribosyltransferase_isoform_X2_[Bombyx_mori]
Full name
Queuine tRNA-ribosyltransferase catalytic subunit 1
+ More
Queuine tRNA-ribosyltransferase
Queuine tRNA-ribosyltransferase
Alternative Name
Guanine insertion enzyme
tRNA-guanine transglycosylase
tRNA-guanine transglycosylase
Location in the cell
PlasmaMembrane Reliability : 2.686
Sequence
CDS
ATGAAAACAACATCAGCAGCTCTACGTTTTAATATATCTACTGAATGTAAAATTACAAAAGCACGCTCTGCAGTTATGAAACTTCCCCATTATGATGTGAAGACACCAGTGTTTATGCCAGTAGGAACCCAGGGTACCATGAAAGGTCTTCTACCAGAACAGCTAGAAAATTTAGATTGTGAAATTATACTAGGAAATACATACCACCTAGGAAATCGACCAGGTACAGATGTTTTAGAAAAAGCTGGTGGCCTCCACAAATTTATGGGTTGGAACAGAGCACTCCTTACAGATTCAGGTGGCTTTCAAATGGTGTCATTACTACAATTAGCGGAAATAACTGAAGAAGGAGTTAAGTTTAAATCACCTTATGATGGCTCTGAAACTATGTTGACACCTGAAAGATCAATTGAAATCCAGAACAGCATAGGAGCTGACATTATCATGCAACTAGATGATGTTGTACAAACAACAAACCAAAACTATGAGAGAAACAAAGAAGCTTCAGAAAGGACCAGCAGATGGCTTGACAGATGTCTAAAGGCACACAAGAGACCCACAGAGCAAAGCATATTTCCTATTGTACAAGGTTTATTAAACAATACCTTAAGAGAACAGAGTGCTAAAGAACACATGACTAAAGATGTAAATGGATTTGCTATTGGAGGGTTAAGTGGGGGTGAAGCAAAGGATGATTTCTGGCCAATGGTTAGCTTGAGCACAGGAATACTTGATAAAAACAAGCCACGATATCTCATGGGTGTTGGAGTTGCCATTGATCTTGTAGTGTGTGTGGCTCTTGGTGTTGACATGTTTGACTGTGTTTTCCCAACTCGTACAGCAAGGTTTGGGTGTGCACTTGTGAATAAAGGACAATTGAGTTTGAAACAAAAAAAATATGAAACAGATCTAAATCCAATTGACAAAGATTGTACTTGTTCAACATGTTTGAATTACACAAGGGCTTATCTCCACAGTATTGTAACAGTAGAAACAGTGTCTTGCCATCTACTTTCTGTGCACAATATTGCTTATCAGATGCGTCTTATGAGGACTATGAGAGAAAATATTGAAAATGGTACATTTCCAGCTTTTGTACAACAATTTGTAAACGAAGTTTATCCCAATGGCCAATATCCTACTTGGGTTGTAAATTCACTTAGCTCAGTTGGAATAAATTTATGCCATCTTTAA
Protein
MKTTSAALRFNISTECKITKARSAVMKLPHYDVKTPVFMPVGTQGTMKGLLPEQLENLDCEIILGNTYHLGNRPGTDVLEKAGGLHKFMGWNRALLTDSGGFQMVSLLQLAEITEEGVKFKSPYDGSETMLTPERSIEIQNSIGADIIMQLDDVVQTTNQNYERNKEASERTSRWLDRCLKAHKRPTEQSIFPIVQGLLNNTLREQSAKEHMTKDVNGFAIGGLSGGEAKDDFWPMVSLSTGILDKNKPRYLMGVGVAIDLVVCVALGVDMFDCVFPTRTARFGCALVNKGQLSLKQKKYETDLNPIDKDCTCSTCLNYTRAYLHSIVTVETVSCHLLSVHNIAYQMRLMRTMRENIENGTFPAFVQQFVNEVYPNGQYPTWVVNSLSSVGINLCHL
Summary
Description
Catalytic subunit of the queuine tRNA-ribosyltransferase (TGT) that catalyzes the base-exchange of a guanine (G) residue with queuine (Q) at position 34 (anticodon wobble position) in tRNAs with GU(N) anticodons (tRNA-Asp, -Asn, -His and -Tyr), resulting in the hypermodified nucleoside queuosine (7-(((4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanosine). Catalysis occurs through a double-displacement mechanism. The nucleophile active site attacks the C1' of nucleotide 34 to detach the guanine base from the RNA, forming a covalent enzyme-RNA intermediate. The proton acceptor active site deprotonates the incoming queuine, allowing a nucleophilic attack on the C1' of the ribose to form the product.
Catalyzes the base-exchange of a guanine (G) residue with the queuine precursor 7-aminomethyl-7-deazaguanine (PreQ1) at position 34 (anticodon wobble position) in tRNAs with GU(N) anticodons (tRNA-Asp, -Asn, -His and -Tyr). Catalysis occurs through a double-displacement mechanism. The nucleophile active site attacks the C1' of nucleotide 34 to detach the guanine base from the RNA, forming a covalent enzyme-RNA intermediate. The proton acceptor active site deprotonates the incoming PreQ1, allowing a nucleophilic attack on the C1' of the ribose to form the product. After dissociation, two additional enzymatic reactions on the tRNA convert PreQ1 to queuine (Q), resulting in the hypermodified nucleoside queuosine (7-(((4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanosine).
Catalyzes the base-exchange of a guanine (G) residue with the queuine precursor 7-aminomethyl-7-deazaguanine (PreQ1) at position 34 (anticodon wobble position) in tRNAs with GU(N) anticodons (tRNA-Asp, -Asn, -His and -Tyr). Catalysis occurs through a double-displacement mechanism. The nucleophile active site attacks the C1' of nucleotide 34 to detach the guanine base from the RNA, forming a covalent enzyme-RNA intermediate. The proton acceptor active site deprotonates the incoming PreQ1, allowing a nucleophilic attack on the C1' of the ribose to form the product. After dissociation, two additional enzymatic reactions on the tRNA convert PreQ1 to queuine (Q), resulting in the hypermodified nucleoside queuosine (7-(((4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanosine).
Catalytic Activity
7-aminomethyl-7-carbaguanine + guanosine(34) in tRNA = 7-aminomethyl-7-carbaguanosine(34) in tRNA + guanine
guanosine(34) in tRNA + queuine = guanine + queuosine(34) in tRNA
guanosine(34) in tRNA + queuine = guanine + queuosine(34) in tRNA
Cofactor
Zn(2+)
Subunit
Heterodimer of a catalytic subunit and an accessory subunit.
Similarity
Belongs to the queuine tRNA-ribosyltransferase family.
Uniprot
A0A2A4JH85
A0A2H1WD50
A0A194R1U2
A0A194PME1
A0A212F0S6
A0A1B6HA51
+ More
E2BAA4 E9JC11 A0A195CF93 H9J9J8 A0A195EZP9 A0A0J7KPY3 A0A232EZ27 A0A067R860 A0A1B6E4B8 A0A195BHF9 K7IMH4 A0A195DMZ5 A0A0C9PHI5 A0A158P378 A0A151WYZ0 A0A026W238 F4W4F1 A0A0L7R4Q3 A0A1D2NMF6 A0A0M9AD08 A0A088ADA0 A0A1Y1M016 A0A1W4WX86 A0A2A3E0V2 E9GFM3 A0A2H8TJL2 A0A2S2PXM6 A0A0P5R5C3 J9KA28 A0A164W331 R4WKC7 A0A2C9JMJ7 E2ALT7 A0A226EJQ1 A0A146LQZ1 A0A210PTU4 A0A146KPN5 A0A023GJN8 G3MPD7 A0A1S4FS82 Q16RD6 A0A0A9XKY3 A0A0T6B3J9 A0A182KXN2 A0A182WVN3 A0A182HDC7 A0A023EQF3 A0A182VE56 A0A0K8SEW7 A0A293LWV9 Q7QAM7 A0A182I9B4 T1IY85 A0A182U7T7 A0A224Z744 A0A131Z8S7 L7M8Y7 A0A131XKH9 A0A182N6C1 T1HLC0 A0A182P902 A0A182KDZ6 R7VA28 A0A224XQI2 A0A0P5MS87 A0A0V0GCG9 N6T4R3 A7SYI3 B0X9L0 A0A182YIA9 A0A182VXJ4 A0A1I8MEN5 K1R475 E0VVJ2 D3PGB1 B4P3G6 A0A1E1X526 A0A1S3H8H3 A0A182M4F3 A0A1S3JXL2 A0A023FAU1 A0A1Q3F514 A0A1Q3F4G9 B4KIK4 A0A0P4VRF9 A0A1B0BP71 B4ID73 B3N8D7 D6WKP4 A0A1A9XQ12 A0A182REW0 B4GQ01 B3MNA9 A0A084VW17
E2BAA4 E9JC11 A0A195CF93 H9J9J8 A0A195EZP9 A0A0J7KPY3 A0A232EZ27 A0A067R860 A0A1B6E4B8 A0A195BHF9 K7IMH4 A0A195DMZ5 A0A0C9PHI5 A0A158P378 A0A151WYZ0 A0A026W238 F4W4F1 A0A0L7R4Q3 A0A1D2NMF6 A0A0M9AD08 A0A088ADA0 A0A1Y1M016 A0A1W4WX86 A0A2A3E0V2 E9GFM3 A0A2H8TJL2 A0A2S2PXM6 A0A0P5R5C3 J9KA28 A0A164W331 R4WKC7 A0A2C9JMJ7 E2ALT7 A0A226EJQ1 A0A146LQZ1 A0A210PTU4 A0A146KPN5 A0A023GJN8 G3MPD7 A0A1S4FS82 Q16RD6 A0A0A9XKY3 A0A0T6B3J9 A0A182KXN2 A0A182WVN3 A0A182HDC7 A0A023EQF3 A0A182VE56 A0A0K8SEW7 A0A293LWV9 Q7QAM7 A0A182I9B4 T1IY85 A0A182U7T7 A0A224Z744 A0A131Z8S7 L7M8Y7 A0A131XKH9 A0A182N6C1 T1HLC0 A0A182P902 A0A182KDZ6 R7VA28 A0A224XQI2 A0A0P5MS87 A0A0V0GCG9 N6T4R3 A7SYI3 B0X9L0 A0A182YIA9 A0A182VXJ4 A0A1I8MEN5 K1R475 E0VVJ2 D3PGB1 B4P3G6 A0A1E1X526 A0A1S3H8H3 A0A182M4F3 A0A1S3JXL2 A0A023FAU1 A0A1Q3F514 A0A1Q3F4G9 B4KIK4 A0A0P4VRF9 A0A1B0BP71 B4ID73 B3N8D7 D6WKP4 A0A1A9XQ12 A0A182REW0 B4GQ01 B3MNA9 A0A084VW17
EC Number
2.4.2.29
Pubmed
26354079
22118469
20798317
21282665
19121390
28648823
+ More
24845553 20075255 21347285 24508170 30249741 21719571 27289101 28004739 21292972 23691247 15562597 26823975 28812685 22216098 17510324 25401762 20966253 26483478 24945155 12364791 14747013 17210077 28797301 26830274 25576852 28049606 23254933 23537049 17615350 25244985 25315136 22992520 20566863 17994087 17550304 28503490 25474469 27129103 18362917 19820115 24438588
24845553 20075255 21347285 24508170 30249741 21719571 27289101 28004739 21292972 23691247 15562597 26823975 28812685 22216098 17510324 25401762 20966253 26483478 24945155 12364791 14747013 17210077 28797301 26830274 25576852 28049606 23254933 23537049 17615350 25244985 25315136 22992520 20566863 17994087 17550304 28503490 25474469 27129103 18362917 19820115 24438588
EMBL
NWSH01001584
PCG70790.1
ODYU01007836
SOQ50957.1
KQ460870
KPJ11753.1
+ More
KQ459601 KPI93904.1 AGBW02011047 OWR47332.1 GECU01036173 JAS71533.1 GL446696 EFN87359.1 GL771642 EFZ09649.1 KQ977935 KYM98713.1 BABH01004978 BABH01004979 BABH01004980 BABH01004981 KQ981905 KYN33591.1 LBMM01004515 KMQ92336.1 NNAY01001612 OXU23428.1 KK852678 KDR18661.1 GEDC01004537 JAS32761.1 KQ976467 KYM84191.1 KQ980713 KYN14248.1 GBYB01000288 JAG70055.1 ADTU01007774 KQ982649 KYQ53108.1 KK107483 QOIP01000001 EZA50132.1 RLU26904.1 GL887532 EGI70846.1 KQ414657 KOC65847.1 LJIJ01000005 ODN06463.1 KQ435698 KOX80753.1 GEZM01048418 GEZM01048417 JAV76577.1 KZ288467 PBC25393.1 GL732542 EFX81780.1 GFXV01002057 MBW13862.1 GGMS01000940 MBY70143.1 GDIQ01114797 JAL36929.1 ABLF02021040 ABLF02037311 LRGB01001348 KZS12907.1 AK418030 BAN21245.1 GL440629 EFN65607.1 LNIX01000003 OXA57679.1 GDHC01009453 JAQ09176.1 NEDP02005494 OWF39930.1 GDHC01020166 JAP98462.1 GBBM01001312 JAC34106.1 JO843738 AEO35355.1 CH477716 EAT36969.1 GBHO01023293 JAG20311.1 LJIG01016090 KRT81645.1 JXUM01001620 KQ560125 KXJ84342.1 GAPW01001781 JAC11817.1 GBRD01013998 JAG51828.1 GFWV01007806 MAA32536.1 AAAB01008888 EAA08866.3 APCN01003250 JH431671 GFPF01011148 MAA22294.1 GEDV01000784 JAP87773.1 GACK01004697 JAA60337.1 GEFH01002550 JAP66031.1 ACPB03006665 AMQN01000633 KB293808 ELU15387.1 GFTR01005716 JAW10710.1 GDIQ01151967 JAK99758.1 GECL01000315 JAP05809.1 APGK01044115 APGK01044116 APGK01044117 KB741020 KB631628 ENN75119.1 ERL84834.1 DS469924 EDO31230.1 DS232541 EDS43179.1 JH816447 EKC28681.1 DS235812 EEB17398.1 BT120667 ADD24307.1 CM000157 EDW87233.1 GFAC01004841 JAT94347.1 AXCM01003824 GBBI01000257 JAC18455.1 GFDL01012426 JAV22619.1 GFDL01012584 JAV22461.1 CH933807 EDW11347.1 GDKW01002128 JAI54467.1 JXJN01017828 CH480829 EDW45499.1 CH954177 EDV57324.1 KQ971342 EFA02997.1 CH479187 EDW39673.1 CH902620 EDV31066.1 ATLV01017375 KE525164 KFB42161.1
KQ459601 KPI93904.1 AGBW02011047 OWR47332.1 GECU01036173 JAS71533.1 GL446696 EFN87359.1 GL771642 EFZ09649.1 KQ977935 KYM98713.1 BABH01004978 BABH01004979 BABH01004980 BABH01004981 KQ981905 KYN33591.1 LBMM01004515 KMQ92336.1 NNAY01001612 OXU23428.1 KK852678 KDR18661.1 GEDC01004537 JAS32761.1 KQ976467 KYM84191.1 KQ980713 KYN14248.1 GBYB01000288 JAG70055.1 ADTU01007774 KQ982649 KYQ53108.1 KK107483 QOIP01000001 EZA50132.1 RLU26904.1 GL887532 EGI70846.1 KQ414657 KOC65847.1 LJIJ01000005 ODN06463.1 KQ435698 KOX80753.1 GEZM01048418 GEZM01048417 JAV76577.1 KZ288467 PBC25393.1 GL732542 EFX81780.1 GFXV01002057 MBW13862.1 GGMS01000940 MBY70143.1 GDIQ01114797 JAL36929.1 ABLF02021040 ABLF02037311 LRGB01001348 KZS12907.1 AK418030 BAN21245.1 GL440629 EFN65607.1 LNIX01000003 OXA57679.1 GDHC01009453 JAQ09176.1 NEDP02005494 OWF39930.1 GDHC01020166 JAP98462.1 GBBM01001312 JAC34106.1 JO843738 AEO35355.1 CH477716 EAT36969.1 GBHO01023293 JAG20311.1 LJIG01016090 KRT81645.1 JXUM01001620 KQ560125 KXJ84342.1 GAPW01001781 JAC11817.1 GBRD01013998 JAG51828.1 GFWV01007806 MAA32536.1 AAAB01008888 EAA08866.3 APCN01003250 JH431671 GFPF01011148 MAA22294.1 GEDV01000784 JAP87773.1 GACK01004697 JAA60337.1 GEFH01002550 JAP66031.1 ACPB03006665 AMQN01000633 KB293808 ELU15387.1 GFTR01005716 JAW10710.1 GDIQ01151967 JAK99758.1 GECL01000315 JAP05809.1 APGK01044115 APGK01044116 APGK01044117 KB741020 KB631628 ENN75119.1 ERL84834.1 DS469924 EDO31230.1 DS232541 EDS43179.1 JH816447 EKC28681.1 DS235812 EEB17398.1 BT120667 ADD24307.1 CM000157 EDW87233.1 GFAC01004841 JAT94347.1 AXCM01003824 GBBI01000257 JAC18455.1 GFDL01012426 JAV22619.1 GFDL01012584 JAV22461.1 CH933807 EDW11347.1 GDKW01002128 JAI54467.1 JXJN01017828 CH480829 EDW45499.1 CH954177 EDV57324.1 KQ971342 EFA02997.1 CH479187 EDW39673.1 CH902620 EDV31066.1 ATLV01017375 KE525164 KFB42161.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000008237
UP000078542
+ More
UP000005204 UP000078541 UP000036403 UP000215335 UP000027135 UP000078540 UP000002358 UP000078492 UP000005205 UP000075809 UP000053097 UP000279307 UP000007755 UP000053825 UP000094527 UP000053105 UP000005203 UP000192223 UP000242457 UP000000305 UP000007819 UP000076858 UP000076420 UP000000311 UP000198287 UP000242188 UP000008820 UP000075882 UP000076407 UP000069940 UP000249989 UP000075903 UP000007062 UP000075840 UP000075902 UP000075884 UP000015103 UP000075885 UP000075881 UP000014760 UP000019118 UP000030742 UP000001593 UP000002320 UP000076408 UP000075920 UP000095301 UP000005408 UP000009046 UP000002282 UP000085678 UP000075883 UP000009192 UP000092460 UP000001292 UP000008711 UP000007266 UP000092443 UP000075900 UP000008744 UP000007801 UP000030765
UP000005204 UP000078541 UP000036403 UP000215335 UP000027135 UP000078540 UP000002358 UP000078492 UP000005205 UP000075809 UP000053097 UP000279307 UP000007755 UP000053825 UP000094527 UP000053105 UP000005203 UP000192223 UP000242457 UP000000305 UP000007819 UP000076858 UP000076420 UP000000311 UP000198287 UP000242188 UP000008820 UP000075882 UP000076407 UP000069940 UP000249989 UP000075903 UP000007062 UP000075840 UP000075902 UP000075884 UP000015103 UP000075885 UP000075881 UP000014760 UP000019118 UP000030742 UP000001593 UP000002320 UP000076408 UP000075920 UP000095301 UP000005408 UP000009046 UP000002282 UP000085678 UP000075883 UP000009192 UP000092460 UP000001292 UP000008711 UP000007266 UP000092443 UP000075900 UP000008744 UP000007801 UP000030765
PRIDE
Pfam
PF01702 TGT
SUPFAM
SSF51713
SSF51713
Gene 3D
ProteinModelPortal
A0A2A4JH85
A0A2H1WD50
A0A194R1U2
A0A194PME1
A0A212F0S6
A0A1B6HA51
+ More
E2BAA4 E9JC11 A0A195CF93 H9J9J8 A0A195EZP9 A0A0J7KPY3 A0A232EZ27 A0A067R860 A0A1B6E4B8 A0A195BHF9 K7IMH4 A0A195DMZ5 A0A0C9PHI5 A0A158P378 A0A151WYZ0 A0A026W238 F4W4F1 A0A0L7R4Q3 A0A1D2NMF6 A0A0M9AD08 A0A088ADA0 A0A1Y1M016 A0A1W4WX86 A0A2A3E0V2 E9GFM3 A0A2H8TJL2 A0A2S2PXM6 A0A0P5R5C3 J9KA28 A0A164W331 R4WKC7 A0A2C9JMJ7 E2ALT7 A0A226EJQ1 A0A146LQZ1 A0A210PTU4 A0A146KPN5 A0A023GJN8 G3MPD7 A0A1S4FS82 Q16RD6 A0A0A9XKY3 A0A0T6B3J9 A0A182KXN2 A0A182WVN3 A0A182HDC7 A0A023EQF3 A0A182VE56 A0A0K8SEW7 A0A293LWV9 Q7QAM7 A0A182I9B4 T1IY85 A0A182U7T7 A0A224Z744 A0A131Z8S7 L7M8Y7 A0A131XKH9 A0A182N6C1 T1HLC0 A0A182P902 A0A182KDZ6 R7VA28 A0A224XQI2 A0A0P5MS87 A0A0V0GCG9 N6T4R3 A7SYI3 B0X9L0 A0A182YIA9 A0A182VXJ4 A0A1I8MEN5 K1R475 E0VVJ2 D3PGB1 B4P3G6 A0A1E1X526 A0A1S3H8H3 A0A182M4F3 A0A1S3JXL2 A0A023FAU1 A0A1Q3F514 A0A1Q3F4G9 B4KIK4 A0A0P4VRF9 A0A1B0BP71 B4ID73 B3N8D7 D6WKP4 A0A1A9XQ12 A0A182REW0 B4GQ01 B3MNA9 A0A084VW17
E2BAA4 E9JC11 A0A195CF93 H9J9J8 A0A195EZP9 A0A0J7KPY3 A0A232EZ27 A0A067R860 A0A1B6E4B8 A0A195BHF9 K7IMH4 A0A195DMZ5 A0A0C9PHI5 A0A158P378 A0A151WYZ0 A0A026W238 F4W4F1 A0A0L7R4Q3 A0A1D2NMF6 A0A0M9AD08 A0A088ADA0 A0A1Y1M016 A0A1W4WX86 A0A2A3E0V2 E9GFM3 A0A2H8TJL2 A0A2S2PXM6 A0A0P5R5C3 J9KA28 A0A164W331 R4WKC7 A0A2C9JMJ7 E2ALT7 A0A226EJQ1 A0A146LQZ1 A0A210PTU4 A0A146KPN5 A0A023GJN8 G3MPD7 A0A1S4FS82 Q16RD6 A0A0A9XKY3 A0A0T6B3J9 A0A182KXN2 A0A182WVN3 A0A182HDC7 A0A023EQF3 A0A182VE56 A0A0K8SEW7 A0A293LWV9 Q7QAM7 A0A182I9B4 T1IY85 A0A182U7T7 A0A224Z744 A0A131Z8S7 L7M8Y7 A0A131XKH9 A0A182N6C1 T1HLC0 A0A182P902 A0A182KDZ6 R7VA28 A0A224XQI2 A0A0P5MS87 A0A0V0GCG9 N6T4R3 A7SYI3 B0X9L0 A0A182YIA9 A0A182VXJ4 A0A1I8MEN5 K1R475 E0VVJ2 D3PGB1 B4P3G6 A0A1E1X526 A0A1S3H8H3 A0A182M4F3 A0A1S3JXL2 A0A023FAU1 A0A1Q3F514 A0A1Q3F4G9 B4KIK4 A0A0P4VRF9 A0A1B0BP71 B4ID73 B3N8D7 D6WKP4 A0A1A9XQ12 A0A182REW0 B4GQ01 B3MNA9 A0A084VW17
PDB
6H45
E-value=7.94508e-134,
Score=1222
Ontologies
GO
Topology
Subcellular location
Cytoplasm
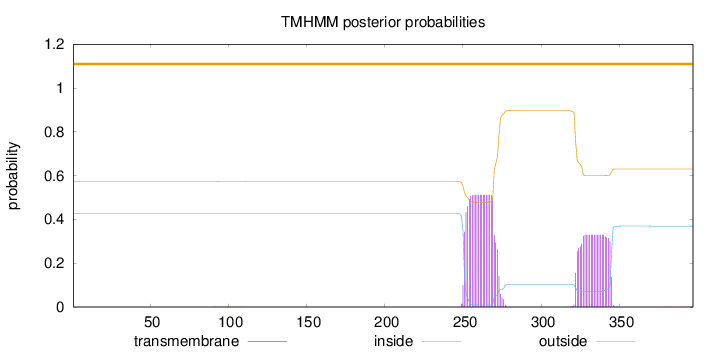
Length:
397
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
17.96535
Exp number, first 60 AAs:
0.00062
Total prob of N-in:
0.42736
outside
1 - 397
Population Genetic Test Statistics
Pi
204.623295
Theta
160.400877
Tajima's D
0.533989
CLR
0.393647
CSRT
0.524723763811809
Interpretation
Uncertain
