Gene
KWMTBOMO01874
Pre Gene Modal
BGIBMGA005953
Annotation
PREDICTED:_alpha-tocopherol_transfer_protein-like_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.339 Cytoskeletal Reliability : 1.554 Nuclear Reliability : 1.674
Sequence
CDS
ATGTCTGTTCGAGCGTTACCTCCGGAGTTAGCAGAAAAAGCCCGCAAAGAGTTAAATGAAGATCCAAGCAGGATTAATAGTGACATTCAACACTTAAAGGACTGGTTGTCCAAACAGCCACATTTACGCGCTAGGACAGACGATCAATGGCTATTATCATTTCTACGAGGATGCAAGTTCAGCTTGGAAAGGACAAAGGAAAAATTAGATTTATATTATTCCGTTCGTACAACTGCACCAGAATTATTCAGTATCAAACCGAGCAATCCATTATTTGATGAAATATTAAGTTTGGGGGCAATATTGGTATTACCTAAGACTGCAACTTCTGATTCTCCACGTGTCAGTATAATCATACCTGGCCGATATGATGCTGATAAGTACAATATTTCCGATGTCATGGCTGTATCTTTAGTCTTACAAAAGATTATTATGATGGAAGACGACAACGCAATGGTTGCCGGCATTTTGTCAATTTTAGATTTAGATACTGTAAAAATAGGTCATTTATTACAAATGACTCCAATGTTCTTGAAAAAAATGGTGGTTGCTTCTCAGGACGCATTACCAGTTCGACTCAAAGGTACCCATTATTTAAATACTCCGACAGGATTCGAAACTGTTTTCAATGCTGTAAAAGCATTACTGAACGAAAAAAATAAGAAACGCCTCTACGTCCACAACCAGAATTATGAAGAGATGTACAAATATATCCCTAAGGACATTTTACCAGTAGAATATGGCGGGAACGGTGGTTCTATTGAAGAAATTATAGATTACTGGAAGAAAAAAGTAAAGGAATACAGTGACTGGTTGGAAGAAGATGATCAATATGGTACGGACGAGTCCAAGCGACCAGGGAAACCGAAGACCGCTGAGGATATGTTCGGGGTCGAGGGATCCTTCAGGCAACTTGAATTCGATTAA
Protein
MSVRALPPELAEKARKELNEDPSRINSDIQHLKDWLSKQPHLRARTDDQWLLSFLRGCKFSLERTKEKLDLYYSVRTTAPELFSIKPSNPLFDEILSLGAILVLPKTATSDSPRVSIIIPGRYDADKYNISDVMAVSLVLQKIIMMEDDNAMVAGILSILDLDTVKIGHLLQMTPMFLKKMVVASQDALPVRLKGTHYLNTPTGFETVFNAVKALLNEKNKKRLYVHNQNYEEMYKYIPKDILPVEYGGNGGSIEEIIDYWKKKVKEYSDWLEEDDQYGTDESKRPGKPKTAEDMFGVEGSFRQLEFD
Summary
Uniprot
H9J8W1
H9J8W2
A0A194R1T7
A0A1E1W1H6
A0A2A4JH87
A0A2H4RMP4
+ More
A0A2H1WW13 A0A194PME6 A0A194PRZ7 A0A194R225 A0A2H4RMN9 A0A212F0Q8 A0A194PKE4 A0A194PLN6 A0A0L7KQJ3 A0A194R7A0 A0A2A4IU45 A0A212F6T3 A0A2A4ISF2 A0A212F6S7 A0A2W1BIJ4 A0A1W7R8C4 A0A1L8DZG8 A0A1L8DZ17 A0A1B0GLZ3 Q16Z99 A0A1B0CUX6 T1E7Q0 Q16ZA0 W5JVC8 A0A182VM76 A0A182KT05 A0A182UMQ1 A0A182KT08 A0A182U0Q4 A0A084WB39 A0A182UHL4 A0A182J0U4 A0A0T6ATJ8 A0A336K545 Q5TMY7 A0A182FW74 A0A182MLC5 A0A182KD53 A0A2H1WW03 A0A182QZB3 W5JX38 A0A182QP18 A0A182PGB9 A0A182HPS1 A0A182X007 A0A194PLP1 A0A194R3C9 A0A182RXC9 A0A182WD92 A0A2H4RMV4 A0A1B0CVS3 A0A182LZL5 A0A182MXC3 A0A2H1WW22 A0A2M4CYG7 A0A1Q3G2T7 A0A182YQ85 A0A2W1BP80 A0A084WB40 A0A2A4IT19 A0A2A4JCS0 A0A0K8TNI0 A0A182IQP8 A0A1J1HGT4 A0A194PKQ7 A0A1J1HK45 B0X6S5 A0A2W1BN19 A0A2H4RMP0 A0A1W4W5W2 A0A0M4ELC6 A0A1E1WUN0 B3M540 B4PD52 A0A336M9G9 A8E768 Q2MGL8 A0A336M2D4 A0A0J9RMX9 A0A2H4RMP2 A0A1E1WRB4 A0A0A1WU57 B4LG89 B3NJG4 B5DR79 B4H5J4 A0A1I8Q731 B4MXY4 A0A034WM90 A0A0T6ATN3 A0A0A1XDP4 A0A3B0K9H1 B3ME36 A0A1L8DZ63
A0A2H1WW13 A0A194PME6 A0A194PRZ7 A0A194R225 A0A2H4RMN9 A0A212F0Q8 A0A194PKE4 A0A194PLN6 A0A0L7KQJ3 A0A194R7A0 A0A2A4IU45 A0A212F6T3 A0A2A4ISF2 A0A212F6S7 A0A2W1BIJ4 A0A1W7R8C4 A0A1L8DZG8 A0A1L8DZ17 A0A1B0GLZ3 Q16Z99 A0A1B0CUX6 T1E7Q0 Q16ZA0 W5JVC8 A0A182VM76 A0A182KT05 A0A182UMQ1 A0A182KT08 A0A182U0Q4 A0A084WB39 A0A182UHL4 A0A182J0U4 A0A0T6ATJ8 A0A336K545 Q5TMY7 A0A182FW74 A0A182MLC5 A0A182KD53 A0A2H1WW03 A0A182QZB3 W5JX38 A0A182QP18 A0A182PGB9 A0A182HPS1 A0A182X007 A0A194PLP1 A0A194R3C9 A0A182RXC9 A0A182WD92 A0A2H4RMV4 A0A1B0CVS3 A0A182LZL5 A0A182MXC3 A0A2H1WW22 A0A2M4CYG7 A0A1Q3G2T7 A0A182YQ85 A0A2W1BP80 A0A084WB40 A0A2A4IT19 A0A2A4JCS0 A0A0K8TNI0 A0A182IQP8 A0A1J1HGT4 A0A194PKQ7 A0A1J1HK45 B0X6S5 A0A2W1BN19 A0A2H4RMP0 A0A1W4W5W2 A0A0M4ELC6 A0A1E1WUN0 B3M540 B4PD52 A0A336M9G9 A8E768 Q2MGL8 A0A336M2D4 A0A0J9RMX9 A0A2H4RMP2 A0A1E1WRB4 A0A0A1WU57 B4LG89 B3NJG4 B5DR79 B4H5J4 A0A1I8Q731 B4MXY4 A0A034WM90 A0A0T6ATN3 A0A0A1XDP4 A0A3B0K9H1 B3ME36 A0A1L8DZ63
Pubmed
EMBL
BABH01004969
BABH01004970
BABH01004971
KQ460870
KPJ11748.1
GDQN01010231
+ More
GDQN01007468 GDQN01002291 GDQN01001042 JAT80823.1 JAT83586.1 JAT88763.1 JAT90012.1 NWSH01001584 PCG70793.1 MG434598 ATY51904.1 ODYU01011491 SOQ57250.1 KQ459601 KPI93909.1 KPI93910.1 KPJ11747.1 MG434597 ATY51903.1 AGBW02011047 OWR47335.1 KPI93911.1 KPI93908.1 JTDY01007407 KOB65224.1 KPJ11746.1 NWSH01007555 PCG62918.1 AGBW02009977 OWR49434.1 PCG62917.1 OWR49433.1 KZ150033 PZC74678.1 GEHC01000255 JAV47390.1 GFDF01002409 JAV11675.1 GFDF01002408 JAV11676.1 AJVK01009188 CH477495 EAT39961.1 AJWK01029765 GAMD01002852 JAA98738.1 EAT39960.1 ADMH02000337 ETN66915.1 ATLV01022312 KE525331 KFB47433.1 LJIG01022828 KRT78512.1 UFQS01000124 UFQT01000124 SSX00096.1 SSX20476.1 AAAB01008986 EAA43295.4 AXCM01009708 AXCM01009709 SOQ57249.1 AXCN02001683 ADMH02000039 ETN68020.1 APCN01003270 KPI93913.1 KPJ11745.1 MG434653 ATY51959.1 AJWK01031086 SOQ57248.1 GGFL01006202 MBW70380.1 GFDL01000931 JAV34114.1 PZC74676.1 KFB47434.1 PCG62915.1 NWSH01002089 PCG69222.1 GDAI01001910 JAI15693.1 CVRI01000002 CRK86620.1 KPI93912.1 CRK86617.1 DS232422 EDS41596.1 PZC74677.1 MG434599 ATY51905.1 CP012525 ALC44235.1 GDQN01000407 JAT90647.1 CH902618 EDV39519.1 CM000159 EDW92800.1 UFQT01000743 SSX26885.1 BT031010 ABV82392.1 AE014296 ABC66124.1 UFQS01000428 UFQT01000428 SSX03855.1 SSX24220.1 CM002912 KMY96769.1 MG434600 ATY51906.1 GDQN01002421 GDQN01002105 GDQN01001504 JAT88633.1 JAT88949.1 JAT89550.1 GBXI01012237 JAD02055.1 CH940647 EDW69397.1 CH954178 EDV50126.2 CH379070 EDY73939.1 CH479211 EDW33046.1 CH963876 EDW76973.1 GAKP01004049 JAC54903.1 KRT78511.1 GBXI01005210 JAD09082.1 OUUW01000009 SPP84770.1 CH902619 EDV35431.1 KPU75643.1 GFDF01002350 JAV11734.1
GDQN01007468 GDQN01002291 GDQN01001042 JAT80823.1 JAT83586.1 JAT88763.1 JAT90012.1 NWSH01001584 PCG70793.1 MG434598 ATY51904.1 ODYU01011491 SOQ57250.1 KQ459601 KPI93909.1 KPI93910.1 KPJ11747.1 MG434597 ATY51903.1 AGBW02011047 OWR47335.1 KPI93911.1 KPI93908.1 JTDY01007407 KOB65224.1 KPJ11746.1 NWSH01007555 PCG62918.1 AGBW02009977 OWR49434.1 PCG62917.1 OWR49433.1 KZ150033 PZC74678.1 GEHC01000255 JAV47390.1 GFDF01002409 JAV11675.1 GFDF01002408 JAV11676.1 AJVK01009188 CH477495 EAT39961.1 AJWK01029765 GAMD01002852 JAA98738.1 EAT39960.1 ADMH02000337 ETN66915.1 ATLV01022312 KE525331 KFB47433.1 LJIG01022828 KRT78512.1 UFQS01000124 UFQT01000124 SSX00096.1 SSX20476.1 AAAB01008986 EAA43295.4 AXCM01009708 AXCM01009709 SOQ57249.1 AXCN02001683 ADMH02000039 ETN68020.1 APCN01003270 KPI93913.1 KPJ11745.1 MG434653 ATY51959.1 AJWK01031086 SOQ57248.1 GGFL01006202 MBW70380.1 GFDL01000931 JAV34114.1 PZC74676.1 KFB47434.1 PCG62915.1 NWSH01002089 PCG69222.1 GDAI01001910 JAI15693.1 CVRI01000002 CRK86620.1 KPI93912.1 CRK86617.1 DS232422 EDS41596.1 PZC74677.1 MG434599 ATY51905.1 CP012525 ALC44235.1 GDQN01000407 JAT90647.1 CH902618 EDV39519.1 CM000159 EDW92800.1 UFQT01000743 SSX26885.1 BT031010 ABV82392.1 AE014296 ABC66124.1 UFQS01000428 UFQT01000428 SSX03855.1 SSX24220.1 CM002912 KMY96769.1 MG434600 ATY51906.1 GDQN01002421 GDQN01002105 GDQN01001504 JAT88633.1 JAT88949.1 JAT89550.1 GBXI01012237 JAD02055.1 CH940647 EDW69397.1 CH954178 EDV50126.2 CH379070 EDY73939.1 CH479211 EDW33046.1 CH963876 EDW76973.1 GAKP01004049 JAC54903.1 KRT78511.1 GBXI01005210 JAD09082.1 OUUW01000009 SPP84770.1 CH902619 EDV35431.1 KPU75643.1 GFDF01002350 JAV11734.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000037510
+ More
UP000092462 UP000008820 UP000092461 UP000000673 UP000075903 UP000075882 UP000075902 UP000030765 UP000075880 UP000007062 UP000069272 UP000075883 UP000075881 UP000075886 UP000075885 UP000075840 UP000076407 UP000075900 UP000075920 UP000075884 UP000076408 UP000183832 UP000002320 UP000192221 UP000092553 UP000007801 UP000002282 UP000000803 UP000008792 UP000008711 UP000001819 UP000008744 UP000095300 UP000007798 UP000268350
UP000092462 UP000008820 UP000092461 UP000000673 UP000075903 UP000075882 UP000075902 UP000030765 UP000075880 UP000007062 UP000069272 UP000075883 UP000075881 UP000075886 UP000075885 UP000075840 UP000076407 UP000075900 UP000075920 UP000075884 UP000076408 UP000183832 UP000002320 UP000192221 UP000092553 UP000007801 UP000002282 UP000000803 UP000008792 UP000008711 UP000001819 UP000008744 UP000095300 UP000007798 UP000268350
Pfam
PF00650 CRAL_TRIO
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J8W1
H9J8W2
A0A194R1T7
A0A1E1W1H6
A0A2A4JH87
A0A2H4RMP4
+ More
A0A2H1WW13 A0A194PME6 A0A194PRZ7 A0A194R225 A0A2H4RMN9 A0A212F0Q8 A0A194PKE4 A0A194PLN6 A0A0L7KQJ3 A0A194R7A0 A0A2A4IU45 A0A212F6T3 A0A2A4ISF2 A0A212F6S7 A0A2W1BIJ4 A0A1W7R8C4 A0A1L8DZG8 A0A1L8DZ17 A0A1B0GLZ3 Q16Z99 A0A1B0CUX6 T1E7Q0 Q16ZA0 W5JVC8 A0A182VM76 A0A182KT05 A0A182UMQ1 A0A182KT08 A0A182U0Q4 A0A084WB39 A0A182UHL4 A0A182J0U4 A0A0T6ATJ8 A0A336K545 Q5TMY7 A0A182FW74 A0A182MLC5 A0A182KD53 A0A2H1WW03 A0A182QZB3 W5JX38 A0A182QP18 A0A182PGB9 A0A182HPS1 A0A182X007 A0A194PLP1 A0A194R3C9 A0A182RXC9 A0A182WD92 A0A2H4RMV4 A0A1B0CVS3 A0A182LZL5 A0A182MXC3 A0A2H1WW22 A0A2M4CYG7 A0A1Q3G2T7 A0A182YQ85 A0A2W1BP80 A0A084WB40 A0A2A4IT19 A0A2A4JCS0 A0A0K8TNI0 A0A182IQP8 A0A1J1HGT4 A0A194PKQ7 A0A1J1HK45 B0X6S5 A0A2W1BN19 A0A2H4RMP0 A0A1W4W5W2 A0A0M4ELC6 A0A1E1WUN0 B3M540 B4PD52 A0A336M9G9 A8E768 Q2MGL8 A0A336M2D4 A0A0J9RMX9 A0A2H4RMP2 A0A1E1WRB4 A0A0A1WU57 B4LG89 B3NJG4 B5DR79 B4H5J4 A0A1I8Q731 B4MXY4 A0A034WM90 A0A0T6ATN3 A0A0A1XDP4 A0A3B0K9H1 B3ME36 A0A1L8DZ63
A0A2H1WW13 A0A194PME6 A0A194PRZ7 A0A194R225 A0A2H4RMN9 A0A212F0Q8 A0A194PKE4 A0A194PLN6 A0A0L7KQJ3 A0A194R7A0 A0A2A4IU45 A0A212F6T3 A0A2A4ISF2 A0A212F6S7 A0A2W1BIJ4 A0A1W7R8C4 A0A1L8DZG8 A0A1L8DZ17 A0A1B0GLZ3 Q16Z99 A0A1B0CUX6 T1E7Q0 Q16ZA0 W5JVC8 A0A182VM76 A0A182KT05 A0A182UMQ1 A0A182KT08 A0A182U0Q4 A0A084WB39 A0A182UHL4 A0A182J0U4 A0A0T6ATJ8 A0A336K545 Q5TMY7 A0A182FW74 A0A182MLC5 A0A182KD53 A0A2H1WW03 A0A182QZB3 W5JX38 A0A182QP18 A0A182PGB9 A0A182HPS1 A0A182X007 A0A194PLP1 A0A194R3C9 A0A182RXC9 A0A182WD92 A0A2H4RMV4 A0A1B0CVS3 A0A182LZL5 A0A182MXC3 A0A2H1WW22 A0A2M4CYG7 A0A1Q3G2T7 A0A182YQ85 A0A2W1BP80 A0A084WB40 A0A2A4IT19 A0A2A4JCS0 A0A0K8TNI0 A0A182IQP8 A0A1J1HGT4 A0A194PKQ7 A0A1J1HK45 B0X6S5 A0A2W1BN19 A0A2H4RMP0 A0A1W4W5W2 A0A0M4ELC6 A0A1E1WUN0 B3M540 B4PD52 A0A336M9G9 A8E768 Q2MGL8 A0A336M2D4 A0A0J9RMX9 A0A2H4RMP2 A0A1E1WRB4 A0A0A1WU57 B4LG89 B3NJG4 B5DR79 B4H5J4 A0A1I8Q731 B4MXY4 A0A034WM90 A0A0T6ATN3 A0A0A1XDP4 A0A3B0K9H1 B3ME36 A0A1L8DZ63
PDB
3W68
E-value=7.63014e-26,
Score=290
Ontologies
GO
Topology
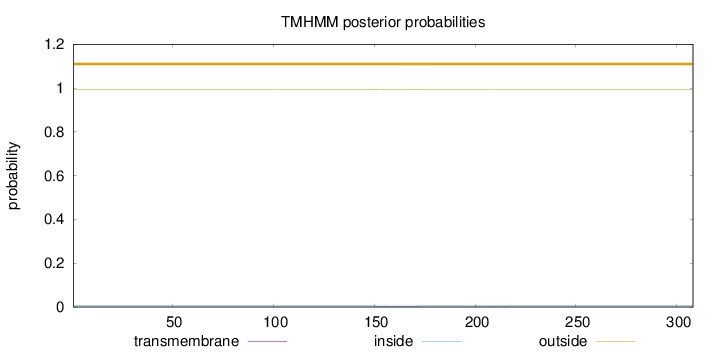
Length:
308
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06562
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00569
outside
1 - 308
Population Genetic Test Statistics
Pi
322.046213
Theta
163.555331
Tajima's D
2.046134
CLR
0.284292
CSRT
0.888555572221389
Interpretation
Uncertain
