Gene
KWMTBOMO01870
Pre Gene Modal
BGIBMGA005957
Annotation
PREDICTED:_uncharacterized_protein_LOC101735801_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.604 Mitochondrial Reliability : 1.888
Sequence
CDS
ATGAAGACCAGAGTACTAAGTCCGGCATTAAGAAAAGTTGCTAAAAACGAACTTGGAGAAGACCTTGAAAAAATCGCAGCAGATCTTATAGTCTTGAGAGCTTGGGTACAAGAATTAGGACAAGGGCAACCAAGTAATCAATGGCTGATAGGTTTCCTACGAGGTAGTAAACACAATGTCGAAAAAGCTAAAAGCAAATTGGAGAAGTATTACAGCTTGAAAGTTTCTATTCCTGAATTTTATACAAATAGAGACCCAATGGACCCGAAAATACAAGCCTTACTGGAACATGGAATATATTTGCCGTTAAGGAAATGTGCCCGTGAAGATTCTCCTCGCCCCGCTATTATCAGAGCAGGCCCACTTTACAAACATCAGCATGCAGTCGCTGATTTAATCAAGATAAGTTTCATGATCGCGGAAATACTGCTTTTGGAAGACGACAACTTCACCGTTGCGGGCAAAGTGAGGCCTCACCGTCACTACTTATAA
Protein
MKTRVLSPALRKVAKNELGEDLEKIAADLIVLRAWVQELGQGQPSNQWLIGFLRGSKHNVEKAKSKLEKYYSLKVSIPEFYTNRDPMDPKIQALLEHGIYLPLRKCAREDSPRPAIIRAGPLYKHQHAVADLIKISFMIAEILLLEDDNFTVAGKVRPHRHYL
Summary
Uniprot
H9J8W5
A0A2A4JCS0
A0A2H1WWA1
A0A2W1BP80
H9J8W4
H9J8W6
+ More
A0A194R3C9 A0A194PLP1 A0A0S2Z370 A0A212FL29 A0A212FL17 A0A2H4RMQ2 A0A182M334 A0A182U4Z5 A0A1J1HK45 A0A1L8DZ58 A0A1L8DZ72 A0A182K803 A0A182IMC1 A0A182GJ38 A0A1S4FJA2 Q16ZA3 A0A084VZS0 A0A182LI81 A0A182TD55 A0A3F2YSV5 Q7Q2Q5 A0A0T6ATJ8 A0A182QUH0 A0A1B0CUX7 A0A0L7LDL8 A0A182KD54 A0A182ULV2 A0A182X1S7 A0A182SKH3 A0A182YI28 A0A1Y1JU40 A0A182SEE1 A0A182VM76 A0A2J7Q750 A0A182KT05 A0A182QZB3 A0A182NNC5 W5JVC8 A0A1J1HFJ3 A0A182FLR5 A0A182YQ84 A0A1J1HGT4 A0A182LZL5 A0A182UHL4 A0A182KD53 A0A0C9QI25 A0A1Y1NBL4 A0A1Y1N9U4 A0A182RXG2 A0A084WB39 Q16Z99 Q5TMY7 Q16ZA0 A0A182MLC5 A0A182WCL5 A0A182X007 W5JS43 A0A182HPS1 A0A336LY11 A0A182UMQ1 A0A182KT08 A0A182PWC3 T1E7Q0 A0A0A1WVX5 A0A1W7R8C4 A0A182WD92 A0A182J0U4 A0A182RXC9 A0A0T6ATN3 A0A1Q3G2T7 A0A1B0EVG2 A0A182U0Q4 B0X6S7 A0A182MXC3 A0A1J1HGF5 A0A1J1HGE9 A0A182PGC0 A0A0A1WLH5 A0A1B0DFH3 A0A182PGB9 A0A336K545 A0A1J1HK53 A0A2H4RMP4 A0A1L8DZ63 W5JUH6 A0A182FW74 A0A182FW75 A0A182QP18 A0A1B0BGD0 A0A1A9UI65 A0A1L8DZH1 A0A0K8TNI0 A0A1B0G0L3 A0A034W6R1
A0A194R3C9 A0A194PLP1 A0A0S2Z370 A0A212FL29 A0A212FL17 A0A2H4RMQ2 A0A182M334 A0A182U4Z5 A0A1J1HK45 A0A1L8DZ58 A0A1L8DZ72 A0A182K803 A0A182IMC1 A0A182GJ38 A0A1S4FJA2 Q16ZA3 A0A084VZS0 A0A182LI81 A0A182TD55 A0A3F2YSV5 Q7Q2Q5 A0A0T6ATJ8 A0A182QUH0 A0A1B0CUX7 A0A0L7LDL8 A0A182KD54 A0A182ULV2 A0A182X1S7 A0A182SKH3 A0A182YI28 A0A1Y1JU40 A0A182SEE1 A0A182VM76 A0A2J7Q750 A0A182KT05 A0A182QZB3 A0A182NNC5 W5JVC8 A0A1J1HFJ3 A0A182FLR5 A0A182YQ84 A0A1J1HGT4 A0A182LZL5 A0A182UHL4 A0A182KD53 A0A0C9QI25 A0A1Y1NBL4 A0A1Y1N9U4 A0A182RXG2 A0A084WB39 Q16Z99 Q5TMY7 Q16ZA0 A0A182MLC5 A0A182WCL5 A0A182X007 W5JS43 A0A182HPS1 A0A336LY11 A0A182UMQ1 A0A182KT08 A0A182PWC3 T1E7Q0 A0A0A1WVX5 A0A1W7R8C4 A0A182WD92 A0A182J0U4 A0A182RXC9 A0A0T6ATN3 A0A1Q3G2T7 A0A1B0EVG2 A0A182U0Q4 B0X6S7 A0A182MXC3 A0A1J1HGF5 A0A1J1HGE9 A0A182PGC0 A0A0A1WLH5 A0A1B0DFH3 A0A182PGB9 A0A336K545 A0A1J1HK53 A0A2H4RMP4 A0A1L8DZ63 W5JUH6 A0A182FW74 A0A182FW75 A0A182QP18 A0A1B0BGD0 A0A1A9UI65 A0A1L8DZH1 A0A0K8TNI0 A0A1B0G0L3 A0A034W6R1
Pubmed
EMBL
BABH01004961
BABH01004962
BABH01004963
BABH01004964
NWSH01002089
PCG69222.1
+ More
ODYU01011491 SOQ57246.1 KZ150033 PZC74676.1 BABH01004965 BABH01004960 KQ460870 KPJ11745.1 KQ459601 KPI93913.1 KT943553 ALQ33311.1 AGBW02007898 OWR54442.1 OWR54443.1 MG434613 ATY51919.1 AXCM01002532 CVRI01000002 CRK86617.1 GFDF01002368 JAV11716.1 GFDF01002367 JAV11717.1 AXCP01008672 JXUM01244789 KQ636530 KXJ62416.1 CH477495 EAT39957.1 ATLV01018955 KE525256 KFB43464.1 APCN01005308 AAAB01008968 EAA13214.4 LJIG01022828 KRT78512.1 AXCN02001905 AJWK01029765 JTDY01001571 KOB73495.1 GEZM01100639 JAV52829.1 NEVH01017447 PNF24412.1 AXCN02001683 ADMH02000337 ETN66915.1 CRK86619.1 CRK86620.1 AXCM01009708 GBYB01003189 JAG72956.1 GEZM01012311 GEZM01012309 JAV92997.1 GEZM01012310 JAV92996.1 ATLV01022312 KE525331 KFB47433.1 EAT39961.1 AAAB01008986 EAA43295.4 EAT39960.1 AXCM01009709 ADMH02000336 ETN66916.1 APCN01003270 UFQS01000278 UFQT01000278 SSX02315.1 SSX22690.1 GAMD01002852 JAA98738.1 GBXI01011709 JAD02583.1 GEHC01000255 JAV47390.1 KRT78511.1 GFDL01000931 JAV34114.1 AJVK01009186 AJVK01009187 DS232422 EDS41598.1 CRK86626.1 CRK86618.1 GBXI01014408 JAC99883.1 AJVK01005789 UFQS01000124 UFQT01000124 SSX00096.1 SSX20476.1 CRK86625.1 MG434598 ATY51904.1 GFDF01002350 JAV11734.1 ETN66918.1 JXJN01013828 GFDF01002402 JAV11682.1 GDAI01001910 JAI15693.1 CCAG010006265 GAKP01007718 GAKP01007717 GAKP01007716 GAKP01007715 JAC51236.1
ODYU01011491 SOQ57246.1 KZ150033 PZC74676.1 BABH01004965 BABH01004960 KQ460870 KPJ11745.1 KQ459601 KPI93913.1 KT943553 ALQ33311.1 AGBW02007898 OWR54442.1 OWR54443.1 MG434613 ATY51919.1 AXCM01002532 CVRI01000002 CRK86617.1 GFDF01002368 JAV11716.1 GFDF01002367 JAV11717.1 AXCP01008672 JXUM01244789 KQ636530 KXJ62416.1 CH477495 EAT39957.1 ATLV01018955 KE525256 KFB43464.1 APCN01005308 AAAB01008968 EAA13214.4 LJIG01022828 KRT78512.1 AXCN02001905 AJWK01029765 JTDY01001571 KOB73495.1 GEZM01100639 JAV52829.1 NEVH01017447 PNF24412.1 AXCN02001683 ADMH02000337 ETN66915.1 CRK86619.1 CRK86620.1 AXCM01009708 GBYB01003189 JAG72956.1 GEZM01012311 GEZM01012309 JAV92997.1 GEZM01012310 JAV92996.1 ATLV01022312 KE525331 KFB47433.1 EAT39961.1 AAAB01008986 EAA43295.4 EAT39960.1 AXCM01009709 ADMH02000336 ETN66916.1 APCN01003270 UFQS01000278 UFQT01000278 SSX02315.1 SSX22690.1 GAMD01002852 JAA98738.1 GBXI01011709 JAD02583.1 GEHC01000255 JAV47390.1 KRT78511.1 GFDL01000931 JAV34114.1 AJVK01009186 AJVK01009187 DS232422 EDS41598.1 CRK86626.1 CRK86618.1 GBXI01014408 JAC99883.1 AJVK01005789 UFQS01000124 UFQT01000124 SSX00096.1 SSX20476.1 CRK86625.1 MG434598 ATY51904.1 GFDF01002350 JAV11734.1 ETN66918.1 JXJN01013828 GFDF01002402 JAV11682.1 GDAI01001910 JAI15693.1 CCAG010006265 GAKP01007718 GAKP01007717 GAKP01007716 GAKP01007715 JAC51236.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000075883
+ More
UP000075902 UP000183832 UP000075881 UP000075880 UP000069940 UP000249989 UP000008820 UP000030765 UP000075882 UP000075840 UP000007062 UP000075886 UP000092461 UP000037510 UP000075903 UP000076407 UP000075901 UP000076408 UP000235965 UP000075884 UP000000673 UP000069272 UP000075900 UP000075920 UP000075885 UP000092462 UP000002320 UP000092460 UP000078200 UP000092444
UP000075902 UP000183832 UP000075881 UP000075880 UP000069940 UP000249989 UP000008820 UP000030765 UP000075882 UP000075840 UP000007062 UP000075886 UP000092461 UP000037510 UP000075903 UP000076407 UP000075901 UP000076408 UP000235965 UP000075884 UP000000673 UP000069272 UP000075900 UP000075920 UP000075885 UP000092462 UP000002320 UP000092460 UP000078200 UP000092444
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J8W5
A0A2A4JCS0
A0A2H1WWA1
A0A2W1BP80
H9J8W4
H9J8W6
+ More
A0A194R3C9 A0A194PLP1 A0A0S2Z370 A0A212FL29 A0A212FL17 A0A2H4RMQ2 A0A182M334 A0A182U4Z5 A0A1J1HK45 A0A1L8DZ58 A0A1L8DZ72 A0A182K803 A0A182IMC1 A0A182GJ38 A0A1S4FJA2 Q16ZA3 A0A084VZS0 A0A182LI81 A0A182TD55 A0A3F2YSV5 Q7Q2Q5 A0A0T6ATJ8 A0A182QUH0 A0A1B0CUX7 A0A0L7LDL8 A0A182KD54 A0A182ULV2 A0A182X1S7 A0A182SKH3 A0A182YI28 A0A1Y1JU40 A0A182SEE1 A0A182VM76 A0A2J7Q750 A0A182KT05 A0A182QZB3 A0A182NNC5 W5JVC8 A0A1J1HFJ3 A0A182FLR5 A0A182YQ84 A0A1J1HGT4 A0A182LZL5 A0A182UHL4 A0A182KD53 A0A0C9QI25 A0A1Y1NBL4 A0A1Y1N9U4 A0A182RXG2 A0A084WB39 Q16Z99 Q5TMY7 Q16ZA0 A0A182MLC5 A0A182WCL5 A0A182X007 W5JS43 A0A182HPS1 A0A336LY11 A0A182UMQ1 A0A182KT08 A0A182PWC3 T1E7Q0 A0A0A1WVX5 A0A1W7R8C4 A0A182WD92 A0A182J0U4 A0A182RXC9 A0A0T6ATN3 A0A1Q3G2T7 A0A1B0EVG2 A0A182U0Q4 B0X6S7 A0A182MXC3 A0A1J1HGF5 A0A1J1HGE9 A0A182PGC0 A0A0A1WLH5 A0A1B0DFH3 A0A182PGB9 A0A336K545 A0A1J1HK53 A0A2H4RMP4 A0A1L8DZ63 W5JUH6 A0A182FW74 A0A182FW75 A0A182QP18 A0A1B0BGD0 A0A1A9UI65 A0A1L8DZH1 A0A0K8TNI0 A0A1B0G0L3 A0A034W6R1
A0A194R3C9 A0A194PLP1 A0A0S2Z370 A0A212FL29 A0A212FL17 A0A2H4RMQ2 A0A182M334 A0A182U4Z5 A0A1J1HK45 A0A1L8DZ58 A0A1L8DZ72 A0A182K803 A0A182IMC1 A0A182GJ38 A0A1S4FJA2 Q16ZA3 A0A084VZS0 A0A182LI81 A0A182TD55 A0A3F2YSV5 Q7Q2Q5 A0A0T6ATJ8 A0A182QUH0 A0A1B0CUX7 A0A0L7LDL8 A0A182KD54 A0A182ULV2 A0A182X1S7 A0A182SKH3 A0A182YI28 A0A1Y1JU40 A0A182SEE1 A0A182VM76 A0A2J7Q750 A0A182KT05 A0A182QZB3 A0A182NNC5 W5JVC8 A0A1J1HFJ3 A0A182FLR5 A0A182YQ84 A0A1J1HGT4 A0A182LZL5 A0A182UHL4 A0A182KD53 A0A0C9QI25 A0A1Y1NBL4 A0A1Y1N9U4 A0A182RXG2 A0A084WB39 Q16Z99 Q5TMY7 Q16ZA0 A0A182MLC5 A0A182WCL5 A0A182X007 W5JS43 A0A182HPS1 A0A336LY11 A0A182UMQ1 A0A182KT08 A0A182PWC3 T1E7Q0 A0A0A1WVX5 A0A1W7R8C4 A0A182WD92 A0A182J0U4 A0A182RXC9 A0A0T6ATN3 A0A1Q3G2T7 A0A1B0EVG2 A0A182U0Q4 B0X6S7 A0A182MXC3 A0A1J1HGF5 A0A1J1HGE9 A0A182PGC0 A0A0A1WLH5 A0A1B0DFH3 A0A182PGB9 A0A336K545 A0A1J1HK53 A0A2H4RMP4 A0A1L8DZ63 W5JUH6 A0A182FW74 A0A182FW75 A0A182QP18 A0A1B0BGD0 A0A1A9UI65 A0A1L8DZH1 A0A0K8TNI0 A0A1B0G0L3 A0A034W6R1
Ontologies
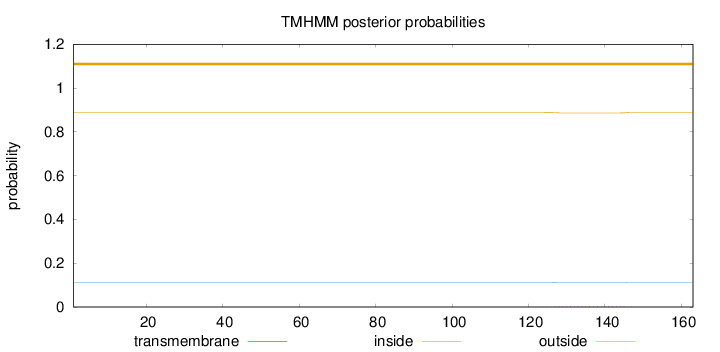
Topology
Length:
163
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05615
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11193
outside
1 - 163
Population Genetic Test Statistics
Pi
225.845346
Theta
172.286573
Tajima's D
1.262153
CLR
0
CSRT
0.728763561821909
Interpretation
Uncertain
