Gene
KWMTBOMO01862
Pre Gene Modal
BGIBMGA005961
Annotation
PREDICTED:_protein_muscleblind_isoform_X5_[Bombyx_mori]
Full name
Protein muscleblind
Alternative Name
Protein mindmelt
Location in the cell
Nuclear Reliability : 3.978
Sequence
CDS
ATGGCCACCATGGTGAACATGAACAGCTTGCTGAACGGCAAGGACTCCCGCTGGTTACAACTCGAAGTATGCAGGGAGTTCCAGCGGAATAAGTGCTCGCGACCAGATACTGAATGCAAATTTGCGCATCCCCCGGCTACCGTCGAGGTGCAGAATGGAAGAGTCACCGCCTGCTACGACAGTATCAAGGGACGGTGTAACCGCGAGAAGCCACCCTGCAAATACTTCCATCCGCCACAACACCTGAAGGATCAGCTGTTGATCAACGGCAGGAATCATCTAGCCCTGAAGAACGCACTAATGCAGCAAATGGGTCTGACACCCGGGCAAGTGTTGCCCGGTCAGGTGCCCGCCGTGGCGACGTCACCTTATTTGTCGGGGGTGCCGGGCGTGGGATCGACGTACGCACAGTACTATGCACCTCAGCTGGTGCCCGCAATGTTGGGTCACGATCCGGCAGCCGCCGCCGCCTCGCCACTCGGTGTCATGCAGCAACCCGTGCTGCAACAGAAATTGCCTCGTACCGATCGTCTCGAGACGGCAGCAGCAGTGCAGTCGCCGGCAACGAGCGCAACTTCAGAGCCGGGCAAGAAACGGGCGGCGCCAACCGAGTTCGAGCCGCCACCGATGGACATGAAGAGCGTCGGTTCCTTCTATTATGATAACTTTGCCTTCCCCGGCGTGGTGCCGTACAAGCGACCGGCTGCCGACAAAGCTGGTGTACCAGTGTATCAGCCGGCGACAACCTACCAGCAACTGATGCAGCTCCAGCAGCCGTTCGTGCCCGTGTCATTTACTGGACACCCTCCCGGTGTGCCAAGATTTTAA
Protein
MATMVNMNSLLNGKDSRWLQLEVCREFQRNKCSRPDTECKFAHPPATVEVQNGRVTACYDSIKGRCNREKPPCKYFHPPQHLKDQLLINGRNHLALKNALMQQMGLTPGQVLPGQVPAVATSPYLSGVPGVGSTYAQYYAPQLVPAMLGHDPAAAAASPLGVMQQPVLQQKLPRTDRLETAAAVQSPATSATSEPGKKRAAPTEFEPPPMDMKSVGSFYYDNFAFPGVVPYKRPAADKAGVPVYQPATTYQQLMQLQQPFVPVSFTGHPPGVPRF
Summary
Description
Required for terminal differentiation of photoreceptor cells. Vital for embryonic development.
Similarity
Belongs to the muscleblind family.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Metal-binding
Nucleus
Reference proteome
Repeat
Sensory transduction
Vision
Zinc
Zinc-finger
Feature
chain Protein muscleblind
splice variant In isoform D.
splice variant In isoform D.
Uniprot
A0A2A4K9M4
A0A1E1WCC8
A0A2W1BDZ1
A0A1Y1N130
V5GUR7
V9I6H0
+ More
A0A1Y1N133 A0A2M4BUL2 A0A0A9YZB9 A0A1B6JIV4 A0A0C9R623 A0A1E1WSN4 A0A0C9RTM5 A0A1B6G9Y7 A0A1B6J5P0 A0A1Y1N142 V5G1U2 A0A1W4X0I9 A0A1W4XA03 A0A0A9YFT7 A0A1B6JIM4 V5GIU1 A0A1Q3EUM2 A0A1B6FIR7 A0A1B6K7X2 A0A069DQ43 A0A1B6DEM7 A0A023F5D2 A0A139WID1 O16011-3 A0A0A9XH65 A0A1B6I4C0 A0A0A9YTS1 B4HMK2 D6WKH9 A0A0C9R3K5 A0A1B6HHG4 A0A1B6ETZ6 A0A1B6JFY6 A0A0C9QWY3 A0A0C9Q974 A0A1B6E1Y4 A0A2M4CIG6 A0A2M4AJ93 A0A2M4BYL1 A0A0C9RCQ2 A0A0A9XVF0 A0A0A9Z6E0 A0A2M4BYB2 A0A336M3I0 A0A1I8P0U0 A0A1B6K5M1 A0A1B6G9S6 A0A2M4BY85 A0A1B6H7V6 A0A1B6H7Y1 A0A336M1F7 A0A0A9YXT6 A0A0C9RMM6 A0A0Q9WGG7 A0A1B6EMV7 A0A0J9U4D6 A0A1B6H9R9 A0A0Q9W3W3 I7BCJ2 A0A0B4K835 A0A1B6ISN7 A0A0Q5VL53 A0A1W4UYR7 A0A0R1DYM3 A0A0J9REM3 A0A0Q9WE84 A0A0J9REV1 I7AZZ5 A0A2H8TPP4 A0A0B4KFA2 A0A0A9ZCA8 A0A336M4U6 A0A0R1DT17 A0A0A9XG83 A0A0L7LDV8 A0A0Q5VVT6 A0A1W4UZ61 A0A0R1DZM0 A0A0P9BZN0 A0A212FPL4 A0A0Q5VN13 A0A336MCX1 J9K0M4 A0A0J9U4D1 A0A2S2QQW3 A0A2S2P312 A0A0R1DT42 A0A0Q9W469 A0A0P8YCS7 A0A0Q9XAB7 A0A0P9BZP1 A0A0Q5VTU5 A0A0Q5VZG9
A0A1Y1N133 A0A2M4BUL2 A0A0A9YZB9 A0A1B6JIV4 A0A0C9R623 A0A1E1WSN4 A0A0C9RTM5 A0A1B6G9Y7 A0A1B6J5P0 A0A1Y1N142 V5G1U2 A0A1W4X0I9 A0A1W4XA03 A0A0A9YFT7 A0A1B6JIM4 V5GIU1 A0A1Q3EUM2 A0A1B6FIR7 A0A1B6K7X2 A0A069DQ43 A0A1B6DEM7 A0A023F5D2 A0A139WID1 O16011-3 A0A0A9XH65 A0A1B6I4C0 A0A0A9YTS1 B4HMK2 D6WKH9 A0A0C9R3K5 A0A1B6HHG4 A0A1B6ETZ6 A0A1B6JFY6 A0A0C9QWY3 A0A0C9Q974 A0A1B6E1Y4 A0A2M4CIG6 A0A2M4AJ93 A0A2M4BYL1 A0A0C9RCQ2 A0A0A9XVF0 A0A0A9Z6E0 A0A2M4BYB2 A0A336M3I0 A0A1I8P0U0 A0A1B6K5M1 A0A1B6G9S6 A0A2M4BY85 A0A1B6H7V6 A0A1B6H7Y1 A0A336M1F7 A0A0A9YXT6 A0A0C9RMM6 A0A0Q9WGG7 A0A1B6EMV7 A0A0J9U4D6 A0A1B6H9R9 A0A0Q9W3W3 I7BCJ2 A0A0B4K835 A0A1B6ISN7 A0A0Q5VL53 A0A1W4UYR7 A0A0R1DYM3 A0A0J9REM3 A0A0Q9WE84 A0A0J9REV1 I7AZZ5 A0A2H8TPP4 A0A0B4KFA2 A0A0A9ZCA8 A0A336M4U6 A0A0R1DT17 A0A0A9XG83 A0A0L7LDV8 A0A0Q5VVT6 A0A1W4UZ61 A0A0R1DZM0 A0A0P9BZN0 A0A212FPL4 A0A0Q5VN13 A0A336MCX1 J9K0M4 A0A0J9U4D1 A0A2S2QQW3 A0A2S2P312 A0A0R1DT42 A0A0Q9W469 A0A0P8YCS7 A0A0Q9XAB7 A0A0P9BZP1 A0A0Q5VTU5 A0A0Q5VZG9
Pubmed
EMBL
NWSH01000026
PCG80598.1
GDQN01006543
GDQN01003717
JAT84511.1
JAT87337.1
+ More
KZ150176 PZC72571.1 GEZM01015475 GEZM01015470 GEZM01015468 GEZM01015467 JAV91623.1 GALX01004513 JAB63953.1 JR035023 AEY56177.1 GEZM01015477 GEZM01015476 GEZM01015473 GEZM01015469 JAV91621.1 GGFJ01007616 MBW56757.1 GBHO01040788 GBHO01040785 GBHO01040777 GBHO01019353 GBHO01007166 JAG02816.1 JAG02819.1 JAG02827.1 JAG24251.1 JAG36438.1 GECU01036862 GECU01021121 GECU01008647 JAS70844.1 JAS86585.1 JAS99059.1 GBYB01008237 JAG78004.1 GDQN01010585 GDQN01001066 JAT80469.1 JAT89988.1 GBYB01010896 JAG80663.1 GECZ01010618 GECZ01010529 GECZ01003874 JAS59151.1 JAS59240.1 JAS65895.1 GECU01017303 GECU01013191 GECU01001860 JAS90403.1 JAS94515.1 JAT05847.1 GEZM01015471 JAV91622.1 GALX01004511 JAB63955.1 GBHO01040789 GBHO01023869 GBHO01018831 GBHO01011672 GBHO01007168 GBHO01004168 GBRD01011723 GBRD01011722 GBRD01007061 GBRD01007060 GBRD01005047 GBRD01000386 GBRD01000384 JAG02815.1 JAG19735.1 JAG24773.1 JAG31932.1 JAG36436.1 JAG39436.1 JAG54101.1 GECU01016782 GECU01015032 GECU01008679 JAS90924.1 JAS92674.1 JAS99027.1 GALX01004512 JAB63954.1 GFDL01016031 JAV19014.1 GECZ01019680 GECZ01015182 GECZ01006198 JAS50089.1 JAS54587.1 JAS63571.1 GECU01018861 GECU01015513 GECU01000175 JAS88845.1 JAS92193.1 JAT07532.1 GBGD01002701 JAC86188.1 GEDC01013144 GEDC01000347 JAS24154.1 JAS36951.1 GBBI01002378 JAC16334.1 KQ971342 KYB27669.1 AF001422 AF001536 AF001625 AF001626 AE013599 GBHO01035699 GBHO01035692 GBHO01035691 GBHO01024603 GBHO01023865 GBHO01019406 JAG07905.1 JAG07912.1 JAG07913.1 JAG19001.1 JAG19739.1 JAG24198.1 GECU01025961 JAS81745.1 GBHO01035694 GBHO01007167 GDHC01018934 JAG07910.1 JAG36437.1 JAP99694.1 CH480816 EDW48270.1 EFA04000.2 GBYB01010899 JAG80666.1 GECU01033566 JAS74140.1 GECZ01028556 GECZ01015774 JAS41213.1 JAS53995.1 GECU01029682 GECU01010278 GECU01009923 JAS78024.1 JAS97428.1 JAS97783.1 GBYB01008239 JAG78006.1 GBYB01010898 JAG80665.1 GEDC01005356 JAS31942.1 GGFL01000946 MBW65124.1 GGFK01007367 MBW40688.1 GGFJ01008961 MBW58102.1 GBYB01010897 JAG80664.1 GBHO01028103 GBHO01018832 JAG15501.1 JAG24772.1 GBHO01040787 GBHO01023870 GBHO01023866 GBHO01007067 GBHO01004165 GBHO01004158 GBRD01007010 GBRD01005046 GBRD01000387 GBRD01000385 JAG02817.1 JAG19734.1 JAG19738.1 JAG36537.1 JAG39439.1 JAG39446.1 JAG58811.1 GGFJ01008934 MBW58075.1 UFQT01000391 SSX23881.1 GECU01000954 JAT06753.1 GECZ01024308 GECZ01010726 JAS45461.1 JAS59043.1 GGFJ01008831 MBW57972.1 GECU01036957 GECU01009538 GECU01004202 JAS70749.1 JAS98168.1 JAT03505.1 GECU01036898 GECU01018306 GECU01002512 JAS70808.1 JAS89400.1 JAT05195.1 SSX23880.1 GBHO01035696 GBHO01024592 GBHO01023863 GBHO01007173 GBHO01007172 GBHO01007171 JAG07908.1 JAG19012.1 JAG19741.1 JAG36431.1 JAG36432.1 JAG36433.1 GBYB01008236 JAG78003.1 CH940648 KRF79568.1 GECZ01030512 JAS39257.1 CM002911 KMY94500.1 GECU01036289 JAS71417.1 KRF79554.1 JN812966 AFO19987.1 AFH08146.1 GECU01027003 GECU01019426 GECU01017762 JAS80703.1 JAS88280.1 JAS89944.1 CH954179 KQS62368.1 CM000158 KRK00362.1 KMY94498.1 KRF79567.1 KMY94502.1 JN812969 AFO19990.1 GFXV01004066 MBW15871.1 AGB93580.1 GBHO01023864 GBHO01004159 GDHC01001195 JAG19740.1 JAG39445.1 JAQ17434.1 SSX23879.1 KRK00358.1 GBHO01040784 GBHO01040783 GBHO01035698 GBHO01024586 GBHO01023867 GBHO01007169 JAG02820.1 JAG02821.1 JAG07906.1 JAG19018.1 JAG19737.1 JAG36435.1 JTDY01001505 KOB73708.1 KQS62363.1 KRK00359.1 CH902619 KPU76770.1 AGBW02001662 OWR55692.1 KQS62364.1 SSX23878.1 ABLF02035697 ABLF02035702 ABLF02051011 KMY94495.1 GGMS01010922 MBY80125.1 GGMR01011230 MBY23849.1 KRK00368.1 KRF79558.1 KPU76769.1 CH933808 KRG05125.1 KPU76779.1 KQS62357.1 KQS62370.1
KZ150176 PZC72571.1 GEZM01015475 GEZM01015470 GEZM01015468 GEZM01015467 JAV91623.1 GALX01004513 JAB63953.1 JR035023 AEY56177.1 GEZM01015477 GEZM01015476 GEZM01015473 GEZM01015469 JAV91621.1 GGFJ01007616 MBW56757.1 GBHO01040788 GBHO01040785 GBHO01040777 GBHO01019353 GBHO01007166 JAG02816.1 JAG02819.1 JAG02827.1 JAG24251.1 JAG36438.1 GECU01036862 GECU01021121 GECU01008647 JAS70844.1 JAS86585.1 JAS99059.1 GBYB01008237 JAG78004.1 GDQN01010585 GDQN01001066 JAT80469.1 JAT89988.1 GBYB01010896 JAG80663.1 GECZ01010618 GECZ01010529 GECZ01003874 JAS59151.1 JAS59240.1 JAS65895.1 GECU01017303 GECU01013191 GECU01001860 JAS90403.1 JAS94515.1 JAT05847.1 GEZM01015471 JAV91622.1 GALX01004511 JAB63955.1 GBHO01040789 GBHO01023869 GBHO01018831 GBHO01011672 GBHO01007168 GBHO01004168 GBRD01011723 GBRD01011722 GBRD01007061 GBRD01007060 GBRD01005047 GBRD01000386 GBRD01000384 JAG02815.1 JAG19735.1 JAG24773.1 JAG31932.1 JAG36436.1 JAG39436.1 JAG54101.1 GECU01016782 GECU01015032 GECU01008679 JAS90924.1 JAS92674.1 JAS99027.1 GALX01004512 JAB63954.1 GFDL01016031 JAV19014.1 GECZ01019680 GECZ01015182 GECZ01006198 JAS50089.1 JAS54587.1 JAS63571.1 GECU01018861 GECU01015513 GECU01000175 JAS88845.1 JAS92193.1 JAT07532.1 GBGD01002701 JAC86188.1 GEDC01013144 GEDC01000347 JAS24154.1 JAS36951.1 GBBI01002378 JAC16334.1 KQ971342 KYB27669.1 AF001422 AF001536 AF001625 AF001626 AE013599 GBHO01035699 GBHO01035692 GBHO01035691 GBHO01024603 GBHO01023865 GBHO01019406 JAG07905.1 JAG07912.1 JAG07913.1 JAG19001.1 JAG19739.1 JAG24198.1 GECU01025961 JAS81745.1 GBHO01035694 GBHO01007167 GDHC01018934 JAG07910.1 JAG36437.1 JAP99694.1 CH480816 EDW48270.1 EFA04000.2 GBYB01010899 JAG80666.1 GECU01033566 JAS74140.1 GECZ01028556 GECZ01015774 JAS41213.1 JAS53995.1 GECU01029682 GECU01010278 GECU01009923 JAS78024.1 JAS97428.1 JAS97783.1 GBYB01008239 JAG78006.1 GBYB01010898 JAG80665.1 GEDC01005356 JAS31942.1 GGFL01000946 MBW65124.1 GGFK01007367 MBW40688.1 GGFJ01008961 MBW58102.1 GBYB01010897 JAG80664.1 GBHO01028103 GBHO01018832 JAG15501.1 JAG24772.1 GBHO01040787 GBHO01023870 GBHO01023866 GBHO01007067 GBHO01004165 GBHO01004158 GBRD01007010 GBRD01005046 GBRD01000387 GBRD01000385 JAG02817.1 JAG19734.1 JAG19738.1 JAG36537.1 JAG39439.1 JAG39446.1 JAG58811.1 GGFJ01008934 MBW58075.1 UFQT01000391 SSX23881.1 GECU01000954 JAT06753.1 GECZ01024308 GECZ01010726 JAS45461.1 JAS59043.1 GGFJ01008831 MBW57972.1 GECU01036957 GECU01009538 GECU01004202 JAS70749.1 JAS98168.1 JAT03505.1 GECU01036898 GECU01018306 GECU01002512 JAS70808.1 JAS89400.1 JAT05195.1 SSX23880.1 GBHO01035696 GBHO01024592 GBHO01023863 GBHO01007173 GBHO01007172 GBHO01007171 JAG07908.1 JAG19012.1 JAG19741.1 JAG36431.1 JAG36432.1 JAG36433.1 GBYB01008236 JAG78003.1 CH940648 KRF79568.1 GECZ01030512 JAS39257.1 CM002911 KMY94500.1 GECU01036289 JAS71417.1 KRF79554.1 JN812966 AFO19987.1 AFH08146.1 GECU01027003 GECU01019426 GECU01017762 JAS80703.1 JAS88280.1 JAS89944.1 CH954179 KQS62368.1 CM000158 KRK00362.1 KMY94498.1 KRF79567.1 KMY94502.1 JN812969 AFO19990.1 GFXV01004066 MBW15871.1 AGB93580.1 GBHO01023864 GBHO01004159 GDHC01001195 JAG19740.1 JAG39445.1 JAQ17434.1 SSX23879.1 KRK00358.1 GBHO01040784 GBHO01040783 GBHO01035698 GBHO01024586 GBHO01023867 GBHO01007169 JAG02820.1 JAG02821.1 JAG07906.1 JAG19018.1 JAG19737.1 JAG36435.1 JTDY01001505 KOB73708.1 KQS62363.1 KRK00359.1 CH902619 KPU76770.1 AGBW02001662 OWR55692.1 KQS62364.1 SSX23878.1 ABLF02035697 ABLF02035702 ABLF02051011 KMY94495.1 GGMS01010922 MBY80125.1 GGMR01011230 MBY23849.1 KRK00368.1 KRF79558.1 KPU76769.1 CH933808 KRG05125.1 KPU76779.1 KQS62357.1 KQS62370.1
Proteomes
PRIDE
Pfam
PF00642 zf-CCCH
Interpro
IPR000571
Znf_CCCH
ProteinModelPortal
A0A2A4K9M4
A0A1E1WCC8
A0A2W1BDZ1
A0A1Y1N130
V5GUR7
V9I6H0
+ More
A0A1Y1N133 A0A2M4BUL2 A0A0A9YZB9 A0A1B6JIV4 A0A0C9R623 A0A1E1WSN4 A0A0C9RTM5 A0A1B6G9Y7 A0A1B6J5P0 A0A1Y1N142 V5G1U2 A0A1W4X0I9 A0A1W4XA03 A0A0A9YFT7 A0A1B6JIM4 V5GIU1 A0A1Q3EUM2 A0A1B6FIR7 A0A1B6K7X2 A0A069DQ43 A0A1B6DEM7 A0A023F5D2 A0A139WID1 O16011-3 A0A0A9XH65 A0A1B6I4C0 A0A0A9YTS1 B4HMK2 D6WKH9 A0A0C9R3K5 A0A1B6HHG4 A0A1B6ETZ6 A0A1B6JFY6 A0A0C9QWY3 A0A0C9Q974 A0A1B6E1Y4 A0A2M4CIG6 A0A2M4AJ93 A0A2M4BYL1 A0A0C9RCQ2 A0A0A9XVF0 A0A0A9Z6E0 A0A2M4BYB2 A0A336M3I0 A0A1I8P0U0 A0A1B6K5M1 A0A1B6G9S6 A0A2M4BY85 A0A1B6H7V6 A0A1B6H7Y1 A0A336M1F7 A0A0A9YXT6 A0A0C9RMM6 A0A0Q9WGG7 A0A1B6EMV7 A0A0J9U4D6 A0A1B6H9R9 A0A0Q9W3W3 I7BCJ2 A0A0B4K835 A0A1B6ISN7 A0A0Q5VL53 A0A1W4UYR7 A0A0R1DYM3 A0A0J9REM3 A0A0Q9WE84 A0A0J9REV1 I7AZZ5 A0A2H8TPP4 A0A0B4KFA2 A0A0A9ZCA8 A0A336M4U6 A0A0R1DT17 A0A0A9XG83 A0A0L7LDV8 A0A0Q5VVT6 A0A1W4UZ61 A0A0R1DZM0 A0A0P9BZN0 A0A212FPL4 A0A0Q5VN13 A0A336MCX1 J9K0M4 A0A0J9U4D1 A0A2S2QQW3 A0A2S2P312 A0A0R1DT42 A0A0Q9W469 A0A0P8YCS7 A0A0Q9XAB7 A0A0P9BZP1 A0A0Q5VTU5 A0A0Q5VZG9
A0A1Y1N133 A0A2M4BUL2 A0A0A9YZB9 A0A1B6JIV4 A0A0C9R623 A0A1E1WSN4 A0A0C9RTM5 A0A1B6G9Y7 A0A1B6J5P0 A0A1Y1N142 V5G1U2 A0A1W4X0I9 A0A1W4XA03 A0A0A9YFT7 A0A1B6JIM4 V5GIU1 A0A1Q3EUM2 A0A1B6FIR7 A0A1B6K7X2 A0A069DQ43 A0A1B6DEM7 A0A023F5D2 A0A139WID1 O16011-3 A0A0A9XH65 A0A1B6I4C0 A0A0A9YTS1 B4HMK2 D6WKH9 A0A0C9R3K5 A0A1B6HHG4 A0A1B6ETZ6 A0A1B6JFY6 A0A0C9QWY3 A0A0C9Q974 A0A1B6E1Y4 A0A2M4CIG6 A0A2M4AJ93 A0A2M4BYL1 A0A0C9RCQ2 A0A0A9XVF0 A0A0A9Z6E0 A0A2M4BYB2 A0A336M3I0 A0A1I8P0U0 A0A1B6K5M1 A0A1B6G9S6 A0A2M4BY85 A0A1B6H7V6 A0A1B6H7Y1 A0A336M1F7 A0A0A9YXT6 A0A0C9RMM6 A0A0Q9WGG7 A0A1B6EMV7 A0A0J9U4D6 A0A1B6H9R9 A0A0Q9W3W3 I7BCJ2 A0A0B4K835 A0A1B6ISN7 A0A0Q5VL53 A0A1W4UYR7 A0A0R1DYM3 A0A0J9REM3 A0A0Q9WE84 A0A0J9REV1 I7AZZ5 A0A2H8TPP4 A0A0B4KFA2 A0A0A9ZCA8 A0A336M4U6 A0A0R1DT17 A0A0A9XG83 A0A0L7LDV8 A0A0Q5VVT6 A0A1W4UZ61 A0A0R1DZM0 A0A0P9BZN0 A0A212FPL4 A0A0Q5VN13 A0A336MCX1 J9K0M4 A0A0J9U4D1 A0A2S2QQW3 A0A2S2P312 A0A0R1DT42 A0A0Q9W469 A0A0P8YCS7 A0A0Q9XAB7 A0A0P9BZP1 A0A0Q5VTU5 A0A0Q5VZG9
PDB
3D2N
E-value=4.21235e-30,
Score=326
Ontologies
GO
GO:0046872
GO:0008168
GO:0007601
GO:0050896
GO:0003676
GO:0046716
GO:0048471
GO:0007517
GO:0010468
GO:0006915
GO:0042052
GO:0001751
GO:0031673
GO:0000381
GO:0001654
GO:0030018
GO:0045924
GO:0005737
GO:0005634
GO:0016021
GO:0005515
GO:0015074
GO:0008270
GO:0043565
GO:0043401
GO:0003677
GO:0003707
GO:0004651
Topology
Subcellular location
Nucleus
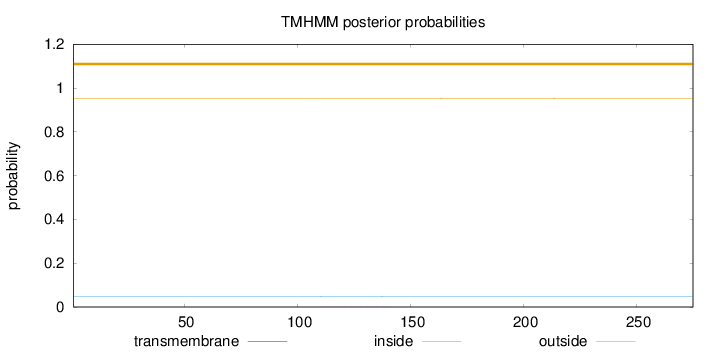
Length:
275
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01372
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04746
outside
1 - 275
Population Genetic Test Statistics
Pi
213.737285
Theta
190.31339
Tajima's D
0.19101
CLR
163.470263
CSRT
0.420628968551572
Interpretation
Uncertain
