Gene
KWMTBOMO01860
Pre Gene Modal
BGIBMGA006189
Annotation
PREDICTED:_long-chain_fatty_acid_transport_protein_4-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.351 Nuclear Reliability : 1.795
Sequence
CDS
ATGGACGCGTTGCTGGCAGCGCTCGTGGCTCTGATGGCAGTTGCCGCCGCGATGGCCGCCGTGCTGAGTACGCTTTCTAAAGCTGCAATATTCGCTATCCTCGCGATAGCGCCCTGTGTGTATCGTTACAGAAAGCAGCTCTACATTATTTTCGTCACTCTTCCACGAGACCTCAAATTCCTATGGCGCTACGCGAATGGCATGGTAAGGTCGAAGCGATGGGGACGTCAAGATGCAACTGTAGCGGAATTGTTCACGCGACGTGCTAAACGGAATCCCGATGCGCCATGTTTTATTGTTGTTGGAGATCGAACGTGGACCTTTGGACAGATTGCGTCGAAATCGAACCAAGTGGCGCGAACTATGCAAGAGCACCTGCAGCTCAAACGCGGTGACGTGGTCTGCGTATTCCTGCCCAACAGCGGCGAGTACATCTGGACGTGGCTTGGCATCGCTAAAGTCGGAGGCGTTTCAGCACTGATCAACAGCAATCTGCGACATAAACCCCTCTTGCACTGCATACAAGTGGCCAATGCAAAGGCTGTAATCTTCTCTGATCAACTTACGGATGCGATTGACGAAATCCGTGATCAACTACCGAAGGGACTAAAAATGTTTCAACTGTATGGAAAGTCTAAACCTGGTATTCTGGATCTTGAGAATGAAATGAGCAAGCATTCAACAGACTACCCAGTCGTCACGGAGAAGCTTCATTATAAAGACACATTGCTTTACATCTACACGTCAGGCACGACGGGAATGCCGAAGGCTGCTGTTATGCCAAACTCCAAGTATTTGTTGATTGTAGCAGCGACGGTCCACATGCTGGGTTTGAATTCATCAGACCGCGTTTACAACTCGCTACCACTGTACCACACTGCGGGCGGTGTGGTGGGAACAGGGGCGGCGCTGGTCGACGGAATCCCCTCTGTGATCAGGCCCAAGTTCTCGGCGAGCAATTTTTGGACTGACTGTATTAAATATGATTGCACGGTGGCCCAGTACATTGGCGAGATGTGTAGATACTTGCTGTCTCAACCGGCGCGACCGACTGATTCGCAGCATCGCGTTCGCATCATGGTCGGCAATGGCATGCGACCCGCTATCTGGCAACAAATAGTGGACCGGTTCAAAATACCGCAGATCAACGAGATCTACGGCGCGACCGAGAGCAATGCAAACATCATCAACGTAGACAATACCTTGGGTGCTGTAGGCTTCCTTCCCAAGCTGGTACCAACCAGCCTTCACCCTATAGCGTTGGTGCGCTCGGACGATCAGGGAAACCTGGTGCGAGGACCTGACGGCTACTGCATCCGATGCCCCCCAAATGAGCCTGGTATGTTCATTGGACTTATCGCCCAGGGGAATGCGTCCCGAGAATATTACGGATACGTCGATAAGAACGACAGCAACAAGAAGTTGGTACGTGACGTTTTCTGTAAGGGAGACGCAGCCTTTGTGAGTGGCGACATCTTAGTGGCAGATGAGTTAGGTTATCTGTTCTTCCGTGATCGCACCGGGGACACGTACAAGTGGAAGGGCGAGAACGTCGCTACCGCCGAGGTCGAGAACGCCATGTCACCAGTGTTGGATGAGAAGGCCTGCGTTGTCTATGGTGTTTCGATCCCTCAGACGGAAGGACGAGTCGGCATGGCTGCCATAGCCGATCCGTTAGGCGAATTGGACTTGAAGAAGCTTGCGCGGGATTTAGATGATTGCTTGCCTTCGTATGCGAGACCAATGTTCCTGCGTTTCATGAATGATTTGGACATAACAAGTACATTCAAACTAAAAAAGTTGCAATACCAAAAGGAAGGTTTTGATCCTGACGTCATCAAGGAGCCGTTATACTTCAGATCCGGAGACAGTTTTGTACCCATAACCTCTCAACTATTCCATGATATTTGTAACGGAAAATTGAAAATTTAA
Protein
MDALLAALVALMAVAAAMAAVLSTLSKAAIFAILAIAPCVYRYRKQLYIIFVTLPRDLKFLWRYANGMVRSKRWGRQDATVAELFTRRAKRNPDAPCFIVVGDRTWTFGQIASKSNQVARTMQEHLQLKRGDVVCVFLPNSGEYIWTWLGIAKVGGVSALINSNLRHKPLLHCIQVANAKAVIFSDQLTDAIDEIRDQLPKGLKMFQLYGKSKPGILDLENEMSKHSTDYPVVTEKLHYKDTLLYIYTSGTTGMPKAAVMPNSKYLLIVAATVHMLGLNSSDRVYNSLPLYHTAGGVVGTGAALVDGIPSVIRPKFSASNFWTDCIKYDCTVAQYIGEMCRYLLSQPARPTDSQHRVRIMVGNGMRPAIWQQIVDRFKIPQINEIYGATESNANIINVDNTLGAVGFLPKLVPTSLHPIALVRSDDQGNLVRGPDGYCIRCPPNEPGMFIGLIAQGNASREYYGYVDKNDSNKKLVRDVFCKGDAAFVSGDILVADELGYLFFRDRTGDTYKWKGENVATAEVENAMSPVLDEKACVVYGVSIPQTEGRVGMAAIADPLGELDLKKLARDLDDCLPSYARPMFLRFMNDLDITSTFKLKKLQYQKEGFDPDVIKEPLYFRSGDSFVPITSQLFHDICNGKLKI
Summary
Uniprot
A0A2A4K8J2
H9J9J6
A0A1V0M8B9
A0A0P0E0E5
A0A076FRK4
A0A194PKR7
+ More
A0A194R3Z1 A0A1L8D6D8 A0A212EVR5 A0A2H1W6H2 A0A232FAF9 A0A0C9S004 K7IPY1 A0A154PK24 A0A088A3U0 A0A023F1R8 A0A0N1ITP6 A0A0L7QWM7 A0A0A9Z8E8 A0A146LH94 A0A0K8TEA3 U5ESE1 A0A0V0G3I3 A0A182GFA7 A0A069DW34 A0A023EVP1 A0A1Q3FPG3 A0A1Q3FZG9 D6WL30 A0A1S4EW84 Q17NU2 A0A0P4VH86 A0A023F4V7 A0A182HFD0 A0A2R7W829 A0A182RNE7 A0A1B6GTZ5 A0A182VYN1 A0A182LYD0 A0A182QWM1 A0A182J0T5 Q7PY63 A0A1S4GCX4 A0A182N6Z6 A0A182U1S3 A0A182KF21 W5J1E2 B0WIY6 A0A151I6U0 A0A182XWA4 T1HML0 A0A0P6CXM8 A0A0C9RBI1 A0A0P5CRG4 A0A162P278 A0A0P5JGV6 A0A0N8ATA2 A0A0P5IY65 A0A0P5K3V0 A0A0P6EVU0 A0A0P5LSW1 A0A0P5K4Z8 A0A0P5G312 A0A034V5Q4 A0A0P5DM29 A0A0P5R8P2 A0A2J7Q856 A0A182FBP1 A0A026WZ46 A0A0A1X4F7 A0A3L8DT65 A0A232F9W6 A0A0P4XYU1 A0A2M3ZA05 K7IPY2 A0A0N7ZQE5 A0A0P4ZDF7 A0A034VIE0 A0A2J7QH75 A0A0P5E1E3 A0A0P4Y375 A0A087ZYN5 A0A0A1XF88 A0A0P5M533 A0A1B0GJK9 W8ATG7 A0A0P5G5D0 A0A1W4XQ51 E2BZR6 A0A0P5MFN0 A0A0P4ZN70 A0A1B6CWE8 B3MJK0 A0A2J7QH78 A0A0P4Y3H5 A0A0P4YUX3 V9IEL8 A0A1B6EP67 E9IX26
A0A194R3Z1 A0A1L8D6D8 A0A212EVR5 A0A2H1W6H2 A0A232FAF9 A0A0C9S004 K7IPY1 A0A154PK24 A0A088A3U0 A0A023F1R8 A0A0N1ITP6 A0A0L7QWM7 A0A0A9Z8E8 A0A146LH94 A0A0K8TEA3 U5ESE1 A0A0V0G3I3 A0A182GFA7 A0A069DW34 A0A023EVP1 A0A1Q3FPG3 A0A1Q3FZG9 D6WL30 A0A1S4EW84 Q17NU2 A0A0P4VH86 A0A023F4V7 A0A182HFD0 A0A2R7W829 A0A182RNE7 A0A1B6GTZ5 A0A182VYN1 A0A182LYD0 A0A182QWM1 A0A182J0T5 Q7PY63 A0A1S4GCX4 A0A182N6Z6 A0A182U1S3 A0A182KF21 W5J1E2 B0WIY6 A0A151I6U0 A0A182XWA4 T1HML0 A0A0P6CXM8 A0A0C9RBI1 A0A0P5CRG4 A0A162P278 A0A0P5JGV6 A0A0N8ATA2 A0A0P5IY65 A0A0P5K3V0 A0A0P6EVU0 A0A0P5LSW1 A0A0P5K4Z8 A0A0P5G312 A0A034V5Q4 A0A0P5DM29 A0A0P5R8P2 A0A2J7Q856 A0A182FBP1 A0A026WZ46 A0A0A1X4F7 A0A3L8DT65 A0A232F9W6 A0A0P4XYU1 A0A2M3ZA05 K7IPY2 A0A0N7ZQE5 A0A0P4ZDF7 A0A034VIE0 A0A2J7QH75 A0A0P5E1E3 A0A0P4Y375 A0A087ZYN5 A0A0A1XF88 A0A0P5M533 A0A1B0GJK9 W8ATG7 A0A0P5G5D0 A0A1W4XQ51 E2BZR6 A0A0P5MFN0 A0A0P4ZN70 A0A1B6CWE8 B3MJK0 A0A2J7QH78 A0A0P4Y3H5 A0A0P4YUX3 V9IEL8 A0A1B6EP67 E9IX26
Pubmed
EMBL
NWSH01000026
PCG80595.1
BABH01004928
BABH01004929
KU755522
ARD71232.1
+ More
KT261737 ALJ30277.1 KF960753 AII21955.1 KQ459601 KPI93922.1 KQ460855 KPJ11960.1 GEYN01000118 JAV02011.1 AGBW02012128 OWR45590.1 ODYU01006649 SOQ48680.1 NNAY01000564 OXU27645.1 GBYB01006136 GBYB01015095 JAG75903.1 JAG84862.1 KQ434943 KZC12231.1 GBBI01003260 JAC15452.1 KQ435759 KOX75694.1 KQ414714 KOC62939.1 GBHO01007187 GBHO01002895 JAG36417.1 JAG40709.1 GDHC01014013 GDHC01011181 JAQ04616.1 JAQ07448.1 GBRD01001918 JAG63903.1 GANO01002398 JAB57473.1 GECL01003465 JAP02659.1 JXUM01001108 JXUM01001109 KQ560116 KXJ84416.1 GBGD01000764 JAC88125.1 GAPW01000350 JAC13248.1 GFDL01005560 JAV29485.1 GFDL01002084 JAV32961.1 KQ971343 EFA03519.2 CH477196 EAT48382.1 GDKW01002815 JAI53780.1 GBBI01002688 JAC16024.1 JXUM01037594 KQ561134 KXJ79609.1 KK854443 PTY15897.1 GECZ01003946 JAS65823.1 AXCM01004450 AXCN02000314 AAAB01008987 EAA01228.5 ADMH02002183 ETN57977.1 DS231954 EDS28847.1 KQ978456 KYM93879.1 ACPB03008862 GDIQ01265532 GDIQ01164679 GDIQ01127246 GDIQ01088141 GDIQ01086151 GDIQ01084605 GDIQ01045818 JAN08586.1 GBYB01004241 GBYB01004242 JAG74008.1 JAG74009.1 GDIP01170573 JAJ52829.1 LRGB01000512 KZS18456.1 GDIQ01202846 JAK48879.1 GDIQ01245044 GDIQ01245043 GDIQ01166265 GDIQ01166263 GDIQ01133981 JAK06682.1 GDIQ01208794 JAK42931.1 GDIQ01190057 JAK61668.1 GDIQ01227702 GDIQ01204648 GDIQ01088142 GDIQ01070047 GDIQ01053515 JAN24690.1 GDIQ01245045 GDIQ01166264 GDIQ01133980 GDIQ01102539 JAK85461.1 GDIQ01201855 GDIQ01189202 GDIQ01189201 JAK62523.1 GDIQ01254338 GDIQ01210254 JAJ97386.1 GAKP01020345 GAKP01020341 JAC38611.1 GDIP01154132 JAJ69270.1 GDIQ01252141 GDIQ01206150 GDIQ01104120 GDIQ01104119 JAL47607.1 NEVH01016978 PNF24770.1 KK107063 EZA61088.1 GBXI01008652 JAD05640.1 QOIP01000005 RLU22958.1 OXU27644.1 GDIP01234973 JAI88428.1 GGFM01004615 MBW25366.1 GDIP01222777 JAJ00625.1 GDIP01214125 JAJ09277.1 GAKP01015886 JAC43066.1 NEVH01013990 PNF27940.1 GDIP01154131 JAJ69271.1 GDIP01233077 JAI90324.1 GBXI01004687 JAD09605.1 GDIQ01160604 JAK91121.1 AJWK01021905 AJWK01021906 AJWK01021907 AJWK01021908 AJWK01021909 AJWK01021910 AJWK01021911 GAMC01014421 GAMC01014420 JAB92135.1 GDIQ01246191 JAK05534.1 GL451657 EFN78828.1 GDIQ01156873 JAK94852.1 GDIP01214126 JAJ09276.1 GEDC01019541 JAS17757.1 CH902620 EDV32368.1 PNF27941.1 GDIP01233078 JAI90323.1 GDIP01222776 JAJ00626.1 JR039957 AEY58921.1 GECZ01030051 JAS39718.1 GL766616 EFZ14911.1
KT261737 ALJ30277.1 KF960753 AII21955.1 KQ459601 KPI93922.1 KQ460855 KPJ11960.1 GEYN01000118 JAV02011.1 AGBW02012128 OWR45590.1 ODYU01006649 SOQ48680.1 NNAY01000564 OXU27645.1 GBYB01006136 GBYB01015095 JAG75903.1 JAG84862.1 KQ434943 KZC12231.1 GBBI01003260 JAC15452.1 KQ435759 KOX75694.1 KQ414714 KOC62939.1 GBHO01007187 GBHO01002895 JAG36417.1 JAG40709.1 GDHC01014013 GDHC01011181 JAQ04616.1 JAQ07448.1 GBRD01001918 JAG63903.1 GANO01002398 JAB57473.1 GECL01003465 JAP02659.1 JXUM01001108 JXUM01001109 KQ560116 KXJ84416.1 GBGD01000764 JAC88125.1 GAPW01000350 JAC13248.1 GFDL01005560 JAV29485.1 GFDL01002084 JAV32961.1 KQ971343 EFA03519.2 CH477196 EAT48382.1 GDKW01002815 JAI53780.1 GBBI01002688 JAC16024.1 JXUM01037594 KQ561134 KXJ79609.1 KK854443 PTY15897.1 GECZ01003946 JAS65823.1 AXCM01004450 AXCN02000314 AAAB01008987 EAA01228.5 ADMH02002183 ETN57977.1 DS231954 EDS28847.1 KQ978456 KYM93879.1 ACPB03008862 GDIQ01265532 GDIQ01164679 GDIQ01127246 GDIQ01088141 GDIQ01086151 GDIQ01084605 GDIQ01045818 JAN08586.1 GBYB01004241 GBYB01004242 JAG74008.1 JAG74009.1 GDIP01170573 JAJ52829.1 LRGB01000512 KZS18456.1 GDIQ01202846 JAK48879.1 GDIQ01245044 GDIQ01245043 GDIQ01166265 GDIQ01166263 GDIQ01133981 JAK06682.1 GDIQ01208794 JAK42931.1 GDIQ01190057 JAK61668.1 GDIQ01227702 GDIQ01204648 GDIQ01088142 GDIQ01070047 GDIQ01053515 JAN24690.1 GDIQ01245045 GDIQ01166264 GDIQ01133980 GDIQ01102539 JAK85461.1 GDIQ01201855 GDIQ01189202 GDIQ01189201 JAK62523.1 GDIQ01254338 GDIQ01210254 JAJ97386.1 GAKP01020345 GAKP01020341 JAC38611.1 GDIP01154132 JAJ69270.1 GDIQ01252141 GDIQ01206150 GDIQ01104120 GDIQ01104119 JAL47607.1 NEVH01016978 PNF24770.1 KK107063 EZA61088.1 GBXI01008652 JAD05640.1 QOIP01000005 RLU22958.1 OXU27644.1 GDIP01234973 JAI88428.1 GGFM01004615 MBW25366.1 GDIP01222777 JAJ00625.1 GDIP01214125 JAJ09277.1 GAKP01015886 JAC43066.1 NEVH01013990 PNF27940.1 GDIP01154131 JAJ69271.1 GDIP01233077 JAI90324.1 GBXI01004687 JAD09605.1 GDIQ01160604 JAK91121.1 AJWK01021905 AJWK01021906 AJWK01021907 AJWK01021908 AJWK01021909 AJWK01021910 AJWK01021911 GAMC01014421 GAMC01014420 JAB92135.1 GDIQ01246191 JAK05534.1 GL451657 EFN78828.1 GDIQ01156873 JAK94852.1 GDIP01214126 JAJ09276.1 GEDC01019541 JAS17757.1 CH902620 EDV32368.1 PNF27941.1 GDIP01233078 JAI90323.1 GDIP01222776 JAJ00626.1 JR039957 AEY58921.1 GECZ01030051 JAS39718.1 GL766616 EFZ14911.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000053240
UP000007151
UP000215335
+ More
UP000002358 UP000076502 UP000005203 UP000053105 UP000053825 UP000069940 UP000249989 UP000007266 UP000008820 UP000075900 UP000075920 UP000075883 UP000075886 UP000075880 UP000007062 UP000075884 UP000075902 UP000075881 UP000000673 UP000002320 UP000078542 UP000076408 UP000015103 UP000076858 UP000235965 UP000069272 UP000053097 UP000279307 UP000092461 UP000192223 UP000008237 UP000007801
UP000002358 UP000076502 UP000005203 UP000053105 UP000053825 UP000069940 UP000249989 UP000007266 UP000008820 UP000075900 UP000075920 UP000075883 UP000075886 UP000075880 UP000007062 UP000075884 UP000075902 UP000075881 UP000000673 UP000002320 UP000078542 UP000076408 UP000015103 UP000076858 UP000235965 UP000069272 UP000053097 UP000279307 UP000092461 UP000192223 UP000008237 UP000007801
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4K8J2
H9J9J6
A0A1V0M8B9
A0A0P0E0E5
A0A076FRK4
A0A194PKR7
+ More
A0A194R3Z1 A0A1L8D6D8 A0A212EVR5 A0A2H1W6H2 A0A232FAF9 A0A0C9S004 K7IPY1 A0A154PK24 A0A088A3U0 A0A023F1R8 A0A0N1ITP6 A0A0L7QWM7 A0A0A9Z8E8 A0A146LH94 A0A0K8TEA3 U5ESE1 A0A0V0G3I3 A0A182GFA7 A0A069DW34 A0A023EVP1 A0A1Q3FPG3 A0A1Q3FZG9 D6WL30 A0A1S4EW84 Q17NU2 A0A0P4VH86 A0A023F4V7 A0A182HFD0 A0A2R7W829 A0A182RNE7 A0A1B6GTZ5 A0A182VYN1 A0A182LYD0 A0A182QWM1 A0A182J0T5 Q7PY63 A0A1S4GCX4 A0A182N6Z6 A0A182U1S3 A0A182KF21 W5J1E2 B0WIY6 A0A151I6U0 A0A182XWA4 T1HML0 A0A0P6CXM8 A0A0C9RBI1 A0A0P5CRG4 A0A162P278 A0A0P5JGV6 A0A0N8ATA2 A0A0P5IY65 A0A0P5K3V0 A0A0P6EVU0 A0A0P5LSW1 A0A0P5K4Z8 A0A0P5G312 A0A034V5Q4 A0A0P5DM29 A0A0P5R8P2 A0A2J7Q856 A0A182FBP1 A0A026WZ46 A0A0A1X4F7 A0A3L8DT65 A0A232F9W6 A0A0P4XYU1 A0A2M3ZA05 K7IPY2 A0A0N7ZQE5 A0A0P4ZDF7 A0A034VIE0 A0A2J7QH75 A0A0P5E1E3 A0A0P4Y375 A0A087ZYN5 A0A0A1XF88 A0A0P5M533 A0A1B0GJK9 W8ATG7 A0A0P5G5D0 A0A1W4XQ51 E2BZR6 A0A0P5MFN0 A0A0P4ZN70 A0A1B6CWE8 B3MJK0 A0A2J7QH78 A0A0P4Y3H5 A0A0P4YUX3 V9IEL8 A0A1B6EP67 E9IX26
A0A194R3Z1 A0A1L8D6D8 A0A212EVR5 A0A2H1W6H2 A0A232FAF9 A0A0C9S004 K7IPY1 A0A154PK24 A0A088A3U0 A0A023F1R8 A0A0N1ITP6 A0A0L7QWM7 A0A0A9Z8E8 A0A146LH94 A0A0K8TEA3 U5ESE1 A0A0V0G3I3 A0A182GFA7 A0A069DW34 A0A023EVP1 A0A1Q3FPG3 A0A1Q3FZG9 D6WL30 A0A1S4EW84 Q17NU2 A0A0P4VH86 A0A023F4V7 A0A182HFD0 A0A2R7W829 A0A182RNE7 A0A1B6GTZ5 A0A182VYN1 A0A182LYD0 A0A182QWM1 A0A182J0T5 Q7PY63 A0A1S4GCX4 A0A182N6Z6 A0A182U1S3 A0A182KF21 W5J1E2 B0WIY6 A0A151I6U0 A0A182XWA4 T1HML0 A0A0P6CXM8 A0A0C9RBI1 A0A0P5CRG4 A0A162P278 A0A0P5JGV6 A0A0N8ATA2 A0A0P5IY65 A0A0P5K3V0 A0A0P6EVU0 A0A0P5LSW1 A0A0P5K4Z8 A0A0P5G312 A0A034V5Q4 A0A0P5DM29 A0A0P5R8P2 A0A2J7Q856 A0A182FBP1 A0A026WZ46 A0A0A1X4F7 A0A3L8DT65 A0A232F9W6 A0A0P4XYU1 A0A2M3ZA05 K7IPY2 A0A0N7ZQE5 A0A0P4ZDF7 A0A034VIE0 A0A2J7QH75 A0A0P5E1E3 A0A0P4Y375 A0A087ZYN5 A0A0A1XF88 A0A0P5M533 A0A1B0GJK9 W8ATG7 A0A0P5G5D0 A0A1W4XQ51 E2BZR6 A0A0P5MFN0 A0A0P4ZN70 A0A1B6CWE8 B3MJK0 A0A2J7QH78 A0A0P4Y3H5 A0A0P4YUX3 V9IEL8 A0A1B6EP67 E9IX26
PDB
3R44
E-value=4.53407e-18,
Score=226
Ontologies
GO
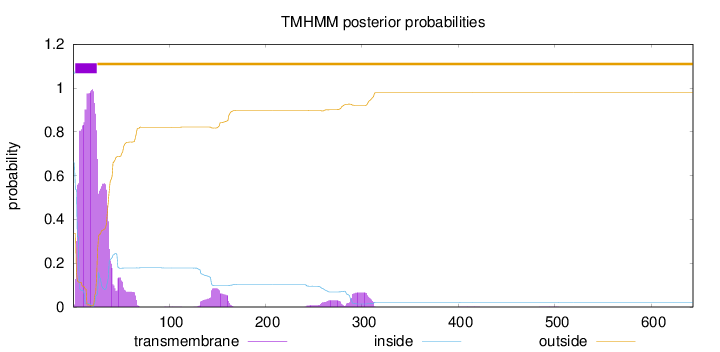
Topology
Length:
643
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
32.1506500000002
Exp number, first 60 AAs:
27.85559
Total prob of N-in:
0.66396
POSSIBLE N-term signal
sequence
inside
1 - 2
TMhelix
3 - 25
outside
26 - 643
Population Genetic Test Statistics
Pi
213.276965
Theta
187.54782
Tajima's D
0.708126
CLR
1.936352
CSRT
0.575671216439178
Interpretation
Uncertain
