Gene
KWMTBOMO01857
Pre Gene Modal
BGIBMGA006185
Annotation
fatty_acid_transport_protein_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.675 Nuclear Reliability : 1.883
Sequence
CDS
ATGAAACCCGGTGCTGTTGACATCGAAAAAAGCAAACAGAACAGTGATTCGGATGGAGTGTCTTGGGGAAAAATATTTTACGTGTTGCTGTCGCTAGCAGTTATCGTGATAGCCTGTGGCGTCGCCTGGGTGTTCCAGGACTGGCTCACCAGTCTTATCATCTTCGTTGTACTTTTGGTGGTATTCACCGTTGGCTATTTCTGGAAATGGCTGTACATCGCGGCCAGAACTGCTCCTAGAGATTTTTCGGCACTGTGGTGTTATGTTAAAATCCTGAGACTTTCGGGTAATTTTGGTAAGAAGAACTGGTCAATGCCCGATATATTCCATGAAAATGTAAAGAGACATCCGAACAAAGCATGCTTCCTTTACGAGAACGAGTCATGGTCCTTTAAACAGGTCGAAGAATTCAGTCTTCGTGTTACGGCAGTGCTTAAGAATCATGGTGTGAAACGTGGAGATGTTGTCGGCGTAATGATGAATAACTGTCCGGAATTGCCGGCGACGTGGCTAGGCGTAGCTCGCATGGGAGGCGTGTCGCCTCTCATAAACACGAACCAAACCGGAAACGCTCTTCTGCATTCGGTTAACGTTGCGAAATGTAACGTTGTGATATACGGAAGCGAGTTTCAGAGTGCATTCGATGAAATATCAAATGAAATAAATCCCGCAATAAAACTGTATAGATACAACCGGAGACCCCTAAATGCATCAGGTGATGCTGTTAGGGTTGTAGAATCGGAAAACGATTTCACTCATATGTTGGAGACTACACCTCCTGCCCCGTGGAGTCTTTCCGACGGAGAAGGCTTCACCGGAAAACTTCTATATATATACACGTCAGGGACTACTGGATTGCCCAAAGCAGCAGTTATATCTCCGTCAAGGATGGTGTTCATGGCATCGGGCGTACATTATTTAGGTGGATTGAGAAAAAACGATATAATGTACTGTCCAATGCCTTTGTATCATTCAGCTGGAGGGTGTATTTCAGTTGGTCAGGCCTTCATATTCGGGTGCACTGTAGCTTTGAGAGCGAAATTTTCGGCGTCTGCATATTTCCCGGATTGCATTAAATTCAAAGCAACGGCCGCTCACTACATTGGGGAGATGTGTCGGTATATCCTAGCAACACCTCCGTCCGCAACTGACAGGCAACACAAAGTTCGAACCGTATACGGCAATGGAATGAGGCCAACCATCTGGACAGAATTCGTCAAACGCTTCAACATCAAACGTGTCGTCGAGTTTTATGGCGCAACAGAAGGCAACGCAAATATCGTAAACATCGACAACAAGACAGGTGCAATAGGATTTGTATCGAGAATCATCCCCGCAGTTTACCCGATAGCGATTCTCAAGGTTGATCAAGAAACTGGCGAACCAATCAGGAATTCCAAAGGACTTTGCCAGCTAGCAAAGCCGTATGAGCCAGGAGTATTTATTGGCAAGATTAAACCTAACAATCCATCAAGGGCGTTCCTGGGTTACGTGGACAAAGAAGCATCTGAGAAGAAAATAGTTAGAGACGTATTTAACATCGGCGATTCTGCGTTTATATCAGGTGACATCCTGGTGGCGGACGAATTAGGTTATCTGTACTTCCGCGACCGCACCGGAGACACGTTCCGGTGGCGCGGAGAGAACGTTTCCACCACAGAGGTCGAGGCGGCCGTGTCTAGATGTGCCAATCAAAGGGACGCTGTAGTCTACGGTGTAGAGATTCCGAATACGGAAGGTCGGGCTGGCATGTGCGGCATAGTGGACATTGAAGGCACATTGGACCTGGATAAACTCGCCAAAGATATTGCGAAAGACCTGCCTAAATACGCGAGGCCAATATTCATTAGAATAATGACCAGCGTTGATATGACAGGAACATTCAAAATGAAGAAAGTGGATTTACAAAAAGAAGGTTATAATCCATCAACTGTGAGCGACAAAATGTTCTTCTTTGAACCAAAACAGAATAAGTACGTGCCCTTAGGAGTTGAAGAATACGAAAAGATTATATCTGGCGCTACGCAGCGCTGCGTTTCGAACTTGCTTTCCACAGGTCGTCGCTCTTCAGTTCGTCGCCTCTCGTTACGAAAGTTAGTCGCGACGATGATAGCCCTTGCCGTCGCCGCCGGTGTGTTCGCAGCCGCTTGGCTCAACATAGGTTTCTATTATTCTCTCACAGCTTCCGTGCTTCTGGTTGTTCTATATTTGATACTCGTTCAGAAAAAATTCTGCTACGTGGCGATAAAAACGATTCCACGGGATGTAAAGGCATTATACACATACATACAGATATTATGGATAACTCGAAAGTTCAGCAAGTTGAATCTCACTGTACCAGATATATTTCATGACACAGTGCAAAAGCATCCAGATAAAGCATGTTTCTTATTCAAGGATGAAGTATGGACATTTAAAGAGAACACCTCAGTTGGCCTGCCCCGGCGTGACACGAATCTGCCAAATGTCATGAGGAAAGCGTCTTTCTAA
Protein
MKPGAVDIEKSKQNSDSDGVSWGKIFYVLLSLAVIVIACGVAWVFQDWLTSLIIFVVLLVVFTVGYFWKWLYIAARTAPRDFSALWCYVKILRLSGNFGKKNWSMPDIFHENVKRHPNKACFLYENESWSFKQVEEFSLRVTAVLKNHGVKRGDVVGVMMNNCPELPATWLGVARMGGVSPLINTNQTGNALLHSVNVAKCNVVIYGSEFQSAFDEISNEINPAIKLYRYNRRPLNASGDAVRVVESENDFTHMLETTPPAPWSLSDGEGFTGKLLYIYTSGTTGLPKAAVISPSRMVFMASGVHYLGGLRKNDIMYCPMPLYHSAGGCISVGQAFIFGCTVALRAKFSASAYFPDCIKFKATAAHYIGEMCRYILATPPSATDRQHKVRTVYGNGMRPTIWTEFVKRFNIKRVVEFYGATEGNANIVNIDNKTGAIGFVSRIIPAVYPIAILKVDQETGEPIRNSKGLCQLAKPYEPGVFIGKIKPNNPSRAFLGYVDKEASEKKIVRDVFNIGDSAFISGDILVADELGYLYFRDRTGDTFRWRGENVSTTEVEAAVSRCANQRDAVVYGVEIPNTEGRAGMCGIVDIEGTLDLDKLAKDIAKDLPKYARPIFIRIMTSVDMTGTFKMKKVDLQKEGYNPSTVSDKMFFFEPKQNKYVPLGVEEYEKIISGATQRCVSNLLSTGRRSSVRRLSLRKLVATMIALAVAAGVFAAAWLNIGFYYSLTASVLLVVLYLILVQKKFCYVAIKTIPRDVKALYTYIQILWITRKFSKLNLTVPDIFHDTVQKHPDKACFLFKDEVWTFKENTSVGLPRRDTNLPNVMRKASF
Summary
Uniprot
B3Y9F5
H9J9J2
A0A2W1BH93
A0A2A4K9F2
E3WCW7
A0A1V0M8C5
+ More
A0A2H1W3D4 A0A0P0EW49 E3WCW8 A0A212EVS6 A0A194PMG1 A0A1L8D6P9 A0A2A4K8S3 A0A0P0DZ69 A0A1V0M8B7 A0A212EVU7 A0A194R363 E2A3N0 D6WL30 A0A151JSV1 A0A151I6U0 A0A1Q3FPG3 A0A1Q3FZG9 A0A195ATB4 A0A023EVP1 A0A158NPT0 A0A182GFA7 T1PEH3 A0A1I8N9H8 A0A1I8N9J3 A0A1B0GJK9 A0A0L0BY49 Q17NU2 A0A1S4EW84 A0A151X8U6 U5ESE1 A0A1I8NPD3 A0A2J7QH75 A0A1L8DYF8 A0A1L8DYP1 A0A1L8DYA9 E2BZR6 B0WIY6 B4LUL2 A0A034V5Q4 A0A2J7QH78 F4W7G4 E9IX26 A0A182KF21 W8ATG7 A0A182VYN1 A0A182J0T5 A0A2A3E8B6 B4MWZ3 A0A0C9RBI1 A0A182LYD0 A0A1W4XQ51 W5J1E2 A0A1S4GCX4 A0A2M3ZA05 Q7PY63 A0A0A1X4F7 A0A182QWM1 A0A182U1S3 A0A182FBP1 A0A182XWA4 A0A182RNE7 A0A1B6CWE8 A0A182HFD0 B3MJK0 A0A182N6Z6 B4KKS4 U4UTR7 A0A232F9W6 A0A3L8DBH0 N6UFM8 A0A026WJP3 K7IPY2 A0A023F4V7 B4JE93 A0A0P4VH86 B4P0Q7 A0A0K8W693 B3N5E1 A0A1A9WHX4 A0A1J1I3Q2 A0A1W4UFG6 A0A034VA16 Q29KS1 B4GSW2 A0A034VAM7 A0A0V0G3I3 A0A1B0BTK8 A0A146M6B7
A0A2H1W3D4 A0A0P0EW49 E3WCW8 A0A212EVS6 A0A194PMG1 A0A1L8D6P9 A0A2A4K8S3 A0A0P0DZ69 A0A1V0M8B7 A0A212EVU7 A0A194R363 E2A3N0 D6WL30 A0A151JSV1 A0A151I6U0 A0A1Q3FPG3 A0A1Q3FZG9 A0A195ATB4 A0A023EVP1 A0A158NPT0 A0A182GFA7 T1PEH3 A0A1I8N9H8 A0A1I8N9J3 A0A1B0GJK9 A0A0L0BY49 Q17NU2 A0A1S4EW84 A0A151X8U6 U5ESE1 A0A1I8NPD3 A0A2J7QH75 A0A1L8DYF8 A0A1L8DYP1 A0A1L8DYA9 E2BZR6 B0WIY6 B4LUL2 A0A034V5Q4 A0A2J7QH78 F4W7G4 E9IX26 A0A182KF21 W8ATG7 A0A182VYN1 A0A182J0T5 A0A2A3E8B6 B4MWZ3 A0A0C9RBI1 A0A182LYD0 A0A1W4XQ51 W5J1E2 A0A1S4GCX4 A0A2M3ZA05 Q7PY63 A0A0A1X4F7 A0A182QWM1 A0A182U1S3 A0A182FBP1 A0A182XWA4 A0A182RNE7 A0A1B6CWE8 A0A182HFD0 B3MJK0 A0A182N6Z6 B4KKS4 U4UTR7 A0A232F9W6 A0A3L8DBH0 N6UFM8 A0A026WJP3 K7IPY2 A0A023F4V7 B4JE93 A0A0P4VH86 B4P0Q7 A0A0K8W693 B3N5E1 A0A1A9WHX4 A0A1J1I3Q2 A0A1W4UFG6 A0A034VA16 Q29KS1 B4GSW2 A0A034VAM7 A0A0V0G3I3 A0A1B0BTK8 A0A146M6B7
Pubmed
19112106
19121390
28756777
20875854
28349297
26445454
+ More
22118469 26354079 20798317 18362917 19820115 24945155 21347285 26483478 25315136 26108605 17510324 17994087 25348373 21719571 21282665 24495485 20920257 23761445 12364791 25830018 25244985 23537049 28648823 30249741 24508170 20075255 25474469 27129103 17550304 18057021 15632085 26823975
22118469 26354079 20798317 18362917 19820115 24945155 21347285 26483478 25315136 26108605 17510324 17994087 25348373 21719571 21282665 24495485 20920257 23761445 12364791 25830018 25244985 23537049 28648823 30249741 24508170 20075255 25474469 27129103 17550304 18057021 15632085 26823975
EMBL
AB451529
BAG68297.1
BABH01004921
BABH01004922
KZ150176
PZC72567.1
+ More
NWSH01000026 PCG80606.1 AB561865 BAJ33523.1 KU755519 ARD71229.1 ODYU01006048 SOQ47558.1 KT261734 ALJ30274.1 AB561866 BAJ33524.1 AGBW02012128 OWR45592.1 KQ459601 KPI93924.1 GEYN01000119 JAV02010.1 PCG80607.1 KT261735 ALJ30275.1 KU755520 ARD71230.1 OWR45591.1 KQ460855 KPJ11959.1 GL436457 EFN71922.1 KQ971343 EFA03519.2 KQ982014 KYN30653.1 KQ978456 KYM93879.1 GFDL01005560 JAV29485.1 GFDL01002084 JAV32961.1 KQ976745 KYM75463.1 GAPW01000350 JAC13248.1 ADTU01022742 ADTU01022743 JXUM01001108 JXUM01001109 KQ560116 KXJ84416.1 KA646525 AFP61154.1 AJWK01021905 AJWK01021906 AJWK01021907 AJWK01021908 AJWK01021909 AJWK01021910 AJWK01021911 JRES01001167 KNC24896.1 CH477196 EAT48382.1 KQ982409 KYQ56708.1 GANO01002398 JAB57473.1 NEVH01013990 PNF27940.1 GFDF01002628 JAV11456.1 GFDF01002506 JAV11578.1 GFDF01002651 JAV11433.1 GL451657 EFN78828.1 DS231954 EDS28847.1 CH940649 EDW64198.1 GAKP01020345 GAKP01020341 JAC38611.1 PNF27941.1 GL887844 EGI69837.1 GL766616 EFZ14911.1 GAMC01014421 GAMC01014420 JAB92135.1 KZ288325 PBC27997.1 CH963857 EDW76632.1 GBYB01004241 GBYB01004242 JAG74008.1 JAG74009.1 AXCM01004450 ADMH02002183 ETN57977.1 AAAB01008987 GGFM01004615 MBW25366.1 EAA01228.5 GBXI01008652 JAD05640.1 AXCN02000314 GEDC01019541 JAS17757.1 JXUM01037594 KQ561134 KXJ79609.1 CH902620 EDV32368.1 CH933807 EDW12738.1 KB632364 ERL93540.1 NNAY01000564 OXU27644.1 QOIP01000011 RLU17248.1 APGK01037735 KB740948 ENN77457.1 KK107183 EZA55891.1 GBBI01002688 JAC16024.1 CH916368 EDW03613.1 GDKW01002815 JAI53780.1 CM000157 EDW87952.1 KRJ97506.1 GDHF01005924 JAI46390.1 CH954177 EDV59020.1 KQS70597.1 CVRI01000036 CRK92993.1 GAKP01020346 GAKP01020342 JAC38610.1 CH379061 EAL33103.3 CH479189 EDW25471.1 GAKP01020344 JAC38608.1 GECL01003465 JAP02659.1 JXJN01020215 GDHC01003326 JAQ15303.1
NWSH01000026 PCG80606.1 AB561865 BAJ33523.1 KU755519 ARD71229.1 ODYU01006048 SOQ47558.1 KT261734 ALJ30274.1 AB561866 BAJ33524.1 AGBW02012128 OWR45592.1 KQ459601 KPI93924.1 GEYN01000119 JAV02010.1 PCG80607.1 KT261735 ALJ30275.1 KU755520 ARD71230.1 OWR45591.1 KQ460855 KPJ11959.1 GL436457 EFN71922.1 KQ971343 EFA03519.2 KQ982014 KYN30653.1 KQ978456 KYM93879.1 GFDL01005560 JAV29485.1 GFDL01002084 JAV32961.1 KQ976745 KYM75463.1 GAPW01000350 JAC13248.1 ADTU01022742 ADTU01022743 JXUM01001108 JXUM01001109 KQ560116 KXJ84416.1 KA646525 AFP61154.1 AJWK01021905 AJWK01021906 AJWK01021907 AJWK01021908 AJWK01021909 AJWK01021910 AJWK01021911 JRES01001167 KNC24896.1 CH477196 EAT48382.1 KQ982409 KYQ56708.1 GANO01002398 JAB57473.1 NEVH01013990 PNF27940.1 GFDF01002628 JAV11456.1 GFDF01002506 JAV11578.1 GFDF01002651 JAV11433.1 GL451657 EFN78828.1 DS231954 EDS28847.1 CH940649 EDW64198.1 GAKP01020345 GAKP01020341 JAC38611.1 PNF27941.1 GL887844 EGI69837.1 GL766616 EFZ14911.1 GAMC01014421 GAMC01014420 JAB92135.1 KZ288325 PBC27997.1 CH963857 EDW76632.1 GBYB01004241 GBYB01004242 JAG74008.1 JAG74009.1 AXCM01004450 ADMH02002183 ETN57977.1 AAAB01008987 GGFM01004615 MBW25366.1 EAA01228.5 GBXI01008652 JAD05640.1 AXCN02000314 GEDC01019541 JAS17757.1 JXUM01037594 KQ561134 KXJ79609.1 CH902620 EDV32368.1 CH933807 EDW12738.1 KB632364 ERL93540.1 NNAY01000564 OXU27644.1 QOIP01000011 RLU17248.1 APGK01037735 KB740948 ENN77457.1 KK107183 EZA55891.1 GBBI01002688 JAC16024.1 CH916368 EDW03613.1 GDKW01002815 JAI53780.1 CM000157 EDW87952.1 KRJ97506.1 GDHF01005924 JAI46390.1 CH954177 EDV59020.1 KQS70597.1 CVRI01000036 CRK92993.1 GAKP01020346 GAKP01020342 JAC38610.1 CH379061 EAL33103.3 CH479189 EDW25471.1 GAKP01020344 JAC38608.1 GECL01003465 JAP02659.1 JXJN01020215 GDHC01003326 JAQ15303.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000000311
+ More
UP000007266 UP000078541 UP000078542 UP000078540 UP000005205 UP000069940 UP000249989 UP000095301 UP000092461 UP000037069 UP000008820 UP000075809 UP000095300 UP000235965 UP000008237 UP000002320 UP000008792 UP000007755 UP000075881 UP000075920 UP000075880 UP000242457 UP000007798 UP000075883 UP000192223 UP000000673 UP000007062 UP000075886 UP000075902 UP000069272 UP000076408 UP000075900 UP000007801 UP000075884 UP000009192 UP000030742 UP000215335 UP000279307 UP000019118 UP000053097 UP000002358 UP000001070 UP000002282 UP000008711 UP000091820 UP000183832 UP000192221 UP000001819 UP000008744 UP000092460
UP000007266 UP000078541 UP000078542 UP000078540 UP000005205 UP000069940 UP000249989 UP000095301 UP000092461 UP000037069 UP000008820 UP000075809 UP000095300 UP000235965 UP000008237 UP000002320 UP000008792 UP000007755 UP000075881 UP000075920 UP000075880 UP000242457 UP000007798 UP000075883 UP000192223 UP000000673 UP000007062 UP000075886 UP000075902 UP000069272 UP000076408 UP000075900 UP000007801 UP000075884 UP000009192 UP000030742 UP000215335 UP000279307 UP000019118 UP000053097 UP000002358 UP000001070 UP000002282 UP000008711 UP000091820 UP000183832 UP000192221 UP000001819 UP000008744 UP000092460
Interpro
Gene 3D
ProteinModelPortal
B3Y9F5
H9J9J2
A0A2W1BH93
A0A2A4K9F2
E3WCW7
A0A1V0M8C5
+ More
A0A2H1W3D4 A0A0P0EW49 E3WCW8 A0A212EVS6 A0A194PMG1 A0A1L8D6P9 A0A2A4K8S3 A0A0P0DZ69 A0A1V0M8B7 A0A212EVU7 A0A194R363 E2A3N0 D6WL30 A0A151JSV1 A0A151I6U0 A0A1Q3FPG3 A0A1Q3FZG9 A0A195ATB4 A0A023EVP1 A0A158NPT0 A0A182GFA7 T1PEH3 A0A1I8N9H8 A0A1I8N9J3 A0A1B0GJK9 A0A0L0BY49 Q17NU2 A0A1S4EW84 A0A151X8U6 U5ESE1 A0A1I8NPD3 A0A2J7QH75 A0A1L8DYF8 A0A1L8DYP1 A0A1L8DYA9 E2BZR6 B0WIY6 B4LUL2 A0A034V5Q4 A0A2J7QH78 F4W7G4 E9IX26 A0A182KF21 W8ATG7 A0A182VYN1 A0A182J0T5 A0A2A3E8B6 B4MWZ3 A0A0C9RBI1 A0A182LYD0 A0A1W4XQ51 W5J1E2 A0A1S4GCX4 A0A2M3ZA05 Q7PY63 A0A0A1X4F7 A0A182QWM1 A0A182U1S3 A0A182FBP1 A0A182XWA4 A0A182RNE7 A0A1B6CWE8 A0A182HFD0 B3MJK0 A0A182N6Z6 B4KKS4 U4UTR7 A0A232F9W6 A0A3L8DBH0 N6UFM8 A0A026WJP3 K7IPY2 A0A023F4V7 B4JE93 A0A0P4VH86 B4P0Q7 A0A0K8W693 B3N5E1 A0A1A9WHX4 A0A1J1I3Q2 A0A1W4UFG6 A0A034VA16 Q29KS1 B4GSW2 A0A034VAM7 A0A0V0G3I3 A0A1B0BTK8 A0A146M6B7
A0A2H1W3D4 A0A0P0EW49 E3WCW8 A0A212EVS6 A0A194PMG1 A0A1L8D6P9 A0A2A4K8S3 A0A0P0DZ69 A0A1V0M8B7 A0A212EVU7 A0A194R363 E2A3N0 D6WL30 A0A151JSV1 A0A151I6U0 A0A1Q3FPG3 A0A1Q3FZG9 A0A195ATB4 A0A023EVP1 A0A158NPT0 A0A182GFA7 T1PEH3 A0A1I8N9H8 A0A1I8N9J3 A0A1B0GJK9 A0A0L0BY49 Q17NU2 A0A1S4EW84 A0A151X8U6 U5ESE1 A0A1I8NPD3 A0A2J7QH75 A0A1L8DYF8 A0A1L8DYP1 A0A1L8DYA9 E2BZR6 B0WIY6 B4LUL2 A0A034V5Q4 A0A2J7QH78 F4W7G4 E9IX26 A0A182KF21 W8ATG7 A0A182VYN1 A0A182J0T5 A0A2A3E8B6 B4MWZ3 A0A0C9RBI1 A0A182LYD0 A0A1W4XQ51 W5J1E2 A0A1S4GCX4 A0A2M3ZA05 Q7PY63 A0A0A1X4F7 A0A182QWM1 A0A182U1S3 A0A182FBP1 A0A182XWA4 A0A182RNE7 A0A1B6CWE8 A0A182HFD0 B3MJK0 A0A182N6Z6 B4KKS4 U4UTR7 A0A232F9W6 A0A3L8DBH0 N6UFM8 A0A026WJP3 K7IPY2 A0A023F4V7 B4JE93 A0A0P4VH86 B4P0Q7 A0A0K8W693 B3N5E1 A0A1A9WHX4 A0A1J1I3Q2 A0A1W4UFG6 A0A034VA16 Q29KS1 B4GSW2 A0A034VAM7 A0A0V0G3I3 A0A1B0BTK8 A0A146M6B7
PDB
3R44
E-value=6.89473e-13,
Score=182
Ontologies
GO
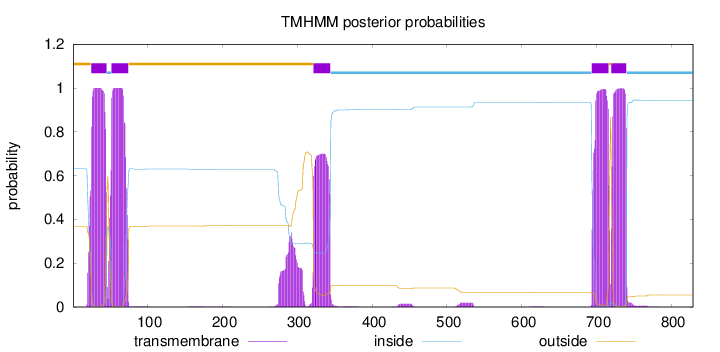
Topology
Length:
829
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
109.81164
Exp number, first 60 AAs:
32.5758
Total prob of N-in:
0.63111
POSSIBLE N-term signal
sequence
outside
1 - 24
TMhelix
25 - 45
inside
46 - 51
TMhelix
52 - 74
outside
75 - 321
TMhelix
322 - 344
inside
345 - 693
TMhelix
694 - 716
outside
717 - 719
TMhelix
720 - 740
inside
741 - 829
Population Genetic Test Statistics
Pi
197.399683
Theta
17.879875
Tajima's D
0.23602
CLR
13.563295
CSRT
0.43632818359082
Interpretation
Uncertain
