Gene
KWMTBOMO01852 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA005966
Annotation
PREDICTED:_glucose-6-phosphate_1-dehydrogenase_[Amyelois_transitella]
Full name
Glucose-6-phosphate 1-dehydrogenase
Alternative Name
Zwischenferment
Location in the cell
Nuclear Reliability : 4.563
Sequence
CDS
ATGGAAAACAAAACTGAATTTGATTATCCGCATACATTTGTACTCCTCGGTGCGTCTGGGGACTTGGCTAGAAAAAAAATTTATCCTACTATCTGGTATTTATATCGTGACAACCTTTTGCCGCATAACACCAAGTTTATTGGTTATGCGCGTACTAAGCAATCTGTTTCCGAAGTAAGAGAAAGGTGTCAAAAGTACATGAAAATACGCCCAGGAGAAGAAGAAAAATTAGAGACATTTTGGAATGCCAATAACTATGTAGCGGGATCTTATGATGGAAGAATAGATTATGAGTTTCTTAACCGAATCATCAGTAATCACGAGAAGGGCCCTATTGCAAACAGAATTTTTTATTTAGCTGTACCTCCAACGGTCTTTGAAGATGTTACTGTAAACATAAAAAATGCCTGTACATCTAGCAAAGGTTTTACTCGTGTTATTATTGAGAAACCTTTTGGACGTGATGATGTCAGCTCTGAGAAATTAAGTAATCACTTAGCTGGACTATTTATCGAAGAACAAATCTACAGAATTGATCATTACCTTGGTAAGGAAATGGTCCAAAACTTGATGACCATAAGATTTGCAAATCAAATATTCAATCCAGCTTGGAATAGAGAAAATATAGCATCTGTTCTTATATCATTTAAAGAACCATTTGGAACTGAAGGTAGAGGAGGGTACTTTGATGATTTTGGCATAATAAGAGATGTCATGCAAAATCATTTACTACAAATACTTTCTTTAGTTGCAATGGAAAAGCCTGTCACTCTAAATCCTAATGATATTAGAGATGAAAAAGTGAAAGTTTTGAGGCATATCCAACCAATAGAATTAAAAGACATTCTAATTGGGCAGTATGTCGGTAATCCTGAAGGTCAAGGTGAAGAAAAATTAGGTTATCTGGATGATCCAACTGTGCCTGGAGACTCCTTAACTCCCACATATGCTCTTGCTACTTTGTGGATCAACAATGCAAGATGGCAAGGTGTCCCCTTTATTCTAAGATGTGGAAAAGCTCTGAATGAAAGAAAAGCAGAAGTTCGTGTTCAGTATAAAGACGTTCCAGGTGATATATTTTCTGGACATGCAAAACGAAATGAACTGGTCATAAGAGTCCAGCCAGGAGAAGCCTTATACTTAAAAATGATGTCTAAATCTCCAGGAATGAAATTTGACTTAGTTGAAACAGAACTAGACCTCACTTATAGTGCACGCTACAAAGAAGCCAGTGTACCTGATGCATATGAAAGGCTCATATTGGATGTGTTCACTGGTACCCAAATGCATTTTGTGCGTAGTGATGAATTAAAAGAAGCATGGCGAGTATTTACACCTATTCTCAAAGATCTCGAAGATAATCACATTAAGCCAGTGCCATATGTTTATGGATCTAGAGGTCCTAAAGAAGCTGATGATAAACTGGCTGGATATGATTTCAAGTATTCTGGGTCTTATAAATGGAAAAAACCTATTCATTGA
Protein
MENKTEFDYPHTFVLLGASGDLARKKIYPTIWYLYRDNLLPHNTKFIGYARTKQSVSEVRERCQKYMKIRPGEEEKLETFWNANNYVAGSYDGRIDYEFLNRIISNHEKGPIANRIFYLAVPPTVFEDVTVNIKNACTSSKGFTRVIIEKPFGRDDVSSEKLSNHLAGLFIEEQIYRIDHYLGKEMVQNLMTIRFANQIFNPAWNRENIASVLISFKEPFGTEGRGGYFDDFGIIRDVMQNHLLQILSLVAMEKPVTLNPNDIRDEKVKVLRHIQPIELKDILIGQYVGNPEGQGEEKLGYLDDPTVPGDSLTPTYALATLWINNARWQGVPFILRCGKALNERKAEVRVQYKDVPGDIFSGHAKRNELVIRVQPGEALYLKMMSKSPGMKFDLVETELDLTYSARYKEASVPDAYERLILDVFTGTQMHFVRSDELKEAWRVFTPILKDLEDNHIKPVPYVYGSRGPKEADDKLAGYDFKYSGSYKWKKPIH
Summary
Description
Catalyzes the rate-limiting step of the oxidative pentose-phosphate pathway, which represents a route for the dissimilation of carbohydrates besides glycolysis.
Catalyzes the rate-limiting step of the oxidative pentose-phosphate pathway, which represents a route for the dissimilation of carbohydrates besides glycolysis. The main function of this enzyme is to provide reducing power (NADPH) and pentose phosphates for fatty acid and nucleic acid synthesis (By similarity).
Catalyzes the rate-limiting step of the oxidative pentose-phosphate pathway, which represents a route for the dissimilation of carbohydrates besides glycolysis. The main function of this enzyme is to provide reducing power (NADPH) and pentose phosphates for fatty acid and nucleic acid synthesis (By similarity).
Catalytic Activity
D-glucose 6-phosphate + NADP(+) = 6-phospho-D-glucono-1,5-lactone + H(+) + NADPH
Similarity
Belongs to the glucose-6-phosphate dehydrogenase family.
Keywords
Carbohydrate metabolism
Glucose metabolism
NADP
Oxidoreductase
Alternative splicing
Complete proteome
Phosphoprotein
Polymorphism
Reference proteome
Feature
chain Glucose-6-phosphate 1-dehydrogenase
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9J8X4
A0A2A4KAC6
A0A2A4K8R3
A0A2S1ZD90
A0A0L7LHT6
A0A194PLQ8
+ More
A0A194R2D9 A0A212F7C7 A0A2H1WSU6 A0A1B1LYT7 D6WKK9 A0A139WIK4 A0A0P6IFH0 A0A164VXB2 A0A067QUT3 A0A1W5YLL4 Q2TLW4 Q2TLW3 A0A0L0CNS3 E9GCL9 A0A0P5FNE9 A0A0P5H692 A0A1I8NG84 A0A1I8NG95 T1PD22 A0A0N8BEX4 V5GYJ0 V5GUY6 B4MSQ8 A0A1Y1KAY8 A0A1I8PHW5 A0A1Y1KEA1 A0A3B0K230 Q29HY8 B4H4R3 A0A0A1XPD3 A0A0A1X5A3 U5EYC9 A0A154P6W4 A0A0R3NZK7 A0A2Z5RDW3 A0A336LTU8 A0A3B0K067 A0A034VUQ7 A0A0K8VCG7 A0A0P5R8T6 A0A0P5XSB4 P41571 A0A182QEB5 A0A0K8UEE3 W8B8T2 A0A084WAQ4 A0A1Q3FEU9 A0A2J7QYS6 A0A2P8Y838 W8ATS3 A0A1D2NGI6 A0A1B6D356 B0WHG8 B7FNK0 A0A2J7QYR4 A0A182FUC7 A0A182J8K4 A0A182P1B9 W5JU82 A0A182WXU3 A0A182NDA4 X2JEF0 A0A182VKK6 A0A182KV52 H2KMF3 A0A0L7QRK3 A0A2M4AU73 A0A226F0P3 B3MQC4 P12646 A0A182Y6T9 A0A0P9A5S1 A0A182WFS6 Q0IEL8 A0A195BFM2 A0A2M4AVT8 P12646-2 A0A182I5W4 A0A1Y9GJ71 A0A182GD35 U4UL56 Q1WKS9 A0A1Y9GIQ5 A0A0U2VLP2 A0A182MVE5 A0A310S6H7 N6TQG0 A0A232F7X7 A0A1J1HGQ8 B3NVS1 Q1WKS8 B4JJG9 A0A158NQV6 A0A0Q5TGW9
A0A194R2D9 A0A212F7C7 A0A2H1WSU6 A0A1B1LYT7 D6WKK9 A0A139WIK4 A0A0P6IFH0 A0A164VXB2 A0A067QUT3 A0A1W5YLL4 Q2TLW4 Q2TLW3 A0A0L0CNS3 E9GCL9 A0A0P5FNE9 A0A0P5H692 A0A1I8NG84 A0A1I8NG95 T1PD22 A0A0N8BEX4 V5GYJ0 V5GUY6 B4MSQ8 A0A1Y1KAY8 A0A1I8PHW5 A0A1Y1KEA1 A0A3B0K230 Q29HY8 B4H4R3 A0A0A1XPD3 A0A0A1X5A3 U5EYC9 A0A154P6W4 A0A0R3NZK7 A0A2Z5RDW3 A0A336LTU8 A0A3B0K067 A0A034VUQ7 A0A0K8VCG7 A0A0P5R8T6 A0A0P5XSB4 P41571 A0A182QEB5 A0A0K8UEE3 W8B8T2 A0A084WAQ4 A0A1Q3FEU9 A0A2J7QYS6 A0A2P8Y838 W8ATS3 A0A1D2NGI6 A0A1B6D356 B0WHG8 B7FNK0 A0A2J7QYR4 A0A182FUC7 A0A182J8K4 A0A182P1B9 W5JU82 A0A182WXU3 A0A182NDA4 X2JEF0 A0A182VKK6 A0A182KV52 H2KMF3 A0A0L7QRK3 A0A2M4AU73 A0A226F0P3 B3MQC4 P12646 A0A182Y6T9 A0A0P9A5S1 A0A182WFS6 Q0IEL8 A0A195BFM2 A0A2M4AVT8 P12646-2 A0A182I5W4 A0A1Y9GJ71 A0A182GD35 U4UL56 Q1WKS9 A0A1Y9GIQ5 A0A0U2VLP2 A0A182MVE5 A0A310S6H7 N6TQG0 A0A232F7X7 A0A1J1HGQ8 B3NVS1 Q1WKS8 B4JJG9 A0A158NQV6 A0A0Q5TGW9
EC Number
1.1.1.49
Pubmed
19121390
26227816
26354079
22118469
27392023
18362917
+ More
19820115 24845553 16469065 26108605 21292972 25315136 17994087 28004739 15632085 25830018 26760975 25348373 8269100 24495485 24438588 29403074 27289101 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20966253 12364791 14747013 17210077 2838391 8913747 12537569 18327897 25244985 17510324 26483478 23537049 16387879 28648823 21347285
19820115 24845553 16469065 26108605 21292972 25315136 17994087 28004739 15632085 25830018 26760975 25348373 8269100 24495485 24438588 29403074 27289101 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20966253 12364791 14747013 17210077 2838391 8913747 12537569 18327897 25244985 17510324 26483478 23537049 16387879 28648823 21347285
EMBL
BABH01004904
NWSH01000026
PCG80602.1
PCG80601.1
MF374345
AWK22771.1
+ More
JTDY01001136 KOB74771.1 KQ459601 KPI93928.1 KQ460855 KPJ11953.1 AGBW02009889 OWR49636.1 ODYU01010805 SOQ56140.1 KU664393 ANS59103.1 KQ971342 EFA03565.2 KYB27681.1 GDIQ01005290 JAN89447.1 LRGB01001361 KZS12751.1 KK852908 KDR13948.1 KX147677 ARI45081.1 AY845218 AAX45784.1 AAX45785.1 JRES01000127 KNC33970.1 GL732539 EFX82581.1 GDIQ01254445 JAJ97279.1 GDIQ01254444 JAJ97280.1 KA646005 AFP60634.1 GDIQ01184467 JAK67258.1 GALX01002988 JAB65478.1 GALX01002989 JAB65477.1 CH963851 EDW75147.2 GEZM01087461 JAV58672.1 GEZM01087462 JAV58671.1 OUUW01000011 SPP86722.1 CH379064 EAL31619.2 CH479209 EDW32675.1 GBXI01001869 JAD12423.1 GBXI01007778 JAD06514.1 GANO01002078 JAB57793.1 KQ434827 KZC07617.1 KRT06404.1 FX983730 BAX07229.1 UFQT01000107 SSX20059.1 SPP86723.1 GAKP01011886 JAC47066.1 GDHF01020138 GDHF01016889 GDHF01015777 GDHF01014827 GDHF01005170 JAI32176.1 JAI35425.1 JAI36537.1 JAI37487.1 JAI47144.1 GDIQ01104232 JAL47494.1 GDIP01068237 JAM35478.1 S67872 AXCN02001783 GDHF01034038 GDHF01027373 JAI18276.1 JAI24941.1 GAMC01016964 GAMC01016962 JAB89591.1 ATLV01022235 KE525330 KFB47298.1 GFDL01008969 JAV26076.1 NEVH01009083 PNF33729.1 PYGN01000816 PSN40423.1 GAMC01016963 JAB89592.1 LJIJ01000056 ODN04076.1 GEDC01017184 JAS20114.1 DS231936 EDS27746.1 BT053690 ACK77607.1 PNF33728.1 ADMH02000433 ETN66505.1 AE014298 AHN59926.1 AAAB01008848 EAL41092.3 KQ414775 KOC61257.1 GGFK01011034 MBW44355.1 LNIX01000001 OXA62506.1 CH902621 EDV44550.1 M26674 M26673 U42738 U42739 U42740 U42741 U42742 U42743 U42744 U42745 U42746 U42747 U42748 U42749 U43165 U43166 U43167 U44721 U45985 AY052079 KPU81635.1 CH477586 EAT38627.1 KQ976500 KYM83009.1 GGFK01011511 MBW44832.1 APCN01003532 JXUM01055187 JXUM01055188 KQ561855 KXJ77310.1 KB632399 ERL94809.1 DQ167797 ABB29562.1 KT279821 ALS35388.1 AXCM01006507 KQ767803 OAD53294.1 APGK01024793 KB740562 ENN80288.1 NNAY01000733 OXU26765.1 CVRI01000003 CRK87059.1 CH954180 EDV46736.1 DQ167798 ABB29563.1 CH916370 EDV99721.1 ADTU01023604 ADTU01023605 KQS30103.1
JTDY01001136 KOB74771.1 KQ459601 KPI93928.1 KQ460855 KPJ11953.1 AGBW02009889 OWR49636.1 ODYU01010805 SOQ56140.1 KU664393 ANS59103.1 KQ971342 EFA03565.2 KYB27681.1 GDIQ01005290 JAN89447.1 LRGB01001361 KZS12751.1 KK852908 KDR13948.1 KX147677 ARI45081.1 AY845218 AAX45784.1 AAX45785.1 JRES01000127 KNC33970.1 GL732539 EFX82581.1 GDIQ01254445 JAJ97279.1 GDIQ01254444 JAJ97280.1 KA646005 AFP60634.1 GDIQ01184467 JAK67258.1 GALX01002988 JAB65478.1 GALX01002989 JAB65477.1 CH963851 EDW75147.2 GEZM01087461 JAV58672.1 GEZM01087462 JAV58671.1 OUUW01000011 SPP86722.1 CH379064 EAL31619.2 CH479209 EDW32675.1 GBXI01001869 JAD12423.1 GBXI01007778 JAD06514.1 GANO01002078 JAB57793.1 KQ434827 KZC07617.1 KRT06404.1 FX983730 BAX07229.1 UFQT01000107 SSX20059.1 SPP86723.1 GAKP01011886 JAC47066.1 GDHF01020138 GDHF01016889 GDHF01015777 GDHF01014827 GDHF01005170 JAI32176.1 JAI35425.1 JAI36537.1 JAI37487.1 JAI47144.1 GDIQ01104232 JAL47494.1 GDIP01068237 JAM35478.1 S67872 AXCN02001783 GDHF01034038 GDHF01027373 JAI18276.1 JAI24941.1 GAMC01016964 GAMC01016962 JAB89591.1 ATLV01022235 KE525330 KFB47298.1 GFDL01008969 JAV26076.1 NEVH01009083 PNF33729.1 PYGN01000816 PSN40423.1 GAMC01016963 JAB89592.1 LJIJ01000056 ODN04076.1 GEDC01017184 JAS20114.1 DS231936 EDS27746.1 BT053690 ACK77607.1 PNF33728.1 ADMH02000433 ETN66505.1 AE014298 AHN59926.1 AAAB01008848 EAL41092.3 KQ414775 KOC61257.1 GGFK01011034 MBW44355.1 LNIX01000001 OXA62506.1 CH902621 EDV44550.1 M26674 M26673 U42738 U42739 U42740 U42741 U42742 U42743 U42744 U42745 U42746 U42747 U42748 U42749 U43165 U43166 U43167 U44721 U45985 AY052079 KPU81635.1 CH477586 EAT38627.1 KQ976500 KYM83009.1 GGFK01011511 MBW44832.1 APCN01003532 JXUM01055187 JXUM01055188 KQ561855 KXJ77310.1 KB632399 ERL94809.1 DQ167797 ABB29562.1 KT279821 ALS35388.1 AXCM01006507 KQ767803 OAD53294.1 APGK01024793 KB740562 ENN80288.1 NNAY01000733 OXU26765.1 CVRI01000003 CRK87059.1 CH954180 EDV46736.1 DQ167798 ABB29563.1 CH916370 EDV99721.1 ADTU01023604 ADTU01023605 KQS30103.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000007151
+ More
UP000007266 UP000076858 UP000027135 UP000037069 UP000000305 UP000095301 UP000007798 UP000095300 UP000268350 UP000001819 UP000008744 UP000076502 UP000075886 UP000030765 UP000235965 UP000245037 UP000094527 UP000002320 UP000069272 UP000075880 UP000075885 UP000000673 UP000076407 UP000075884 UP000000803 UP000075903 UP000075882 UP000007062 UP000053825 UP000198287 UP000007801 UP000076408 UP000075920 UP000008820 UP000078540 UP000075840 UP000069940 UP000249989 UP000030742 UP000075883 UP000019118 UP000215335 UP000183832 UP000008711 UP000001070 UP000005205
UP000007266 UP000076858 UP000027135 UP000037069 UP000000305 UP000095301 UP000007798 UP000095300 UP000268350 UP000001819 UP000008744 UP000076502 UP000075886 UP000030765 UP000235965 UP000245037 UP000094527 UP000002320 UP000069272 UP000075880 UP000075885 UP000000673 UP000076407 UP000075884 UP000000803 UP000075903 UP000075882 UP000007062 UP000053825 UP000198287 UP000007801 UP000076408 UP000075920 UP000008820 UP000078540 UP000075840 UP000069940 UP000249989 UP000030742 UP000075883 UP000019118 UP000215335 UP000183832 UP000008711 UP000001070 UP000005205
Interpro
Gene 3D
ProteinModelPortal
H9J8X4
A0A2A4KAC6
A0A2A4K8R3
A0A2S1ZD90
A0A0L7LHT6
A0A194PLQ8
+ More
A0A194R2D9 A0A212F7C7 A0A2H1WSU6 A0A1B1LYT7 D6WKK9 A0A139WIK4 A0A0P6IFH0 A0A164VXB2 A0A067QUT3 A0A1W5YLL4 Q2TLW4 Q2TLW3 A0A0L0CNS3 E9GCL9 A0A0P5FNE9 A0A0P5H692 A0A1I8NG84 A0A1I8NG95 T1PD22 A0A0N8BEX4 V5GYJ0 V5GUY6 B4MSQ8 A0A1Y1KAY8 A0A1I8PHW5 A0A1Y1KEA1 A0A3B0K230 Q29HY8 B4H4R3 A0A0A1XPD3 A0A0A1X5A3 U5EYC9 A0A154P6W4 A0A0R3NZK7 A0A2Z5RDW3 A0A336LTU8 A0A3B0K067 A0A034VUQ7 A0A0K8VCG7 A0A0P5R8T6 A0A0P5XSB4 P41571 A0A182QEB5 A0A0K8UEE3 W8B8T2 A0A084WAQ4 A0A1Q3FEU9 A0A2J7QYS6 A0A2P8Y838 W8ATS3 A0A1D2NGI6 A0A1B6D356 B0WHG8 B7FNK0 A0A2J7QYR4 A0A182FUC7 A0A182J8K4 A0A182P1B9 W5JU82 A0A182WXU3 A0A182NDA4 X2JEF0 A0A182VKK6 A0A182KV52 H2KMF3 A0A0L7QRK3 A0A2M4AU73 A0A226F0P3 B3MQC4 P12646 A0A182Y6T9 A0A0P9A5S1 A0A182WFS6 Q0IEL8 A0A195BFM2 A0A2M4AVT8 P12646-2 A0A182I5W4 A0A1Y9GJ71 A0A182GD35 U4UL56 Q1WKS9 A0A1Y9GIQ5 A0A0U2VLP2 A0A182MVE5 A0A310S6H7 N6TQG0 A0A232F7X7 A0A1J1HGQ8 B3NVS1 Q1WKS8 B4JJG9 A0A158NQV6 A0A0Q5TGW9
A0A194R2D9 A0A212F7C7 A0A2H1WSU6 A0A1B1LYT7 D6WKK9 A0A139WIK4 A0A0P6IFH0 A0A164VXB2 A0A067QUT3 A0A1W5YLL4 Q2TLW4 Q2TLW3 A0A0L0CNS3 E9GCL9 A0A0P5FNE9 A0A0P5H692 A0A1I8NG84 A0A1I8NG95 T1PD22 A0A0N8BEX4 V5GYJ0 V5GUY6 B4MSQ8 A0A1Y1KAY8 A0A1I8PHW5 A0A1Y1KEA1 A0A3B0K230 Q29HY8 B4H4R3 A0A0A1XPD3 A0A0A1X5A3 U5EYC9 A0A154P6W4 A0A0R3NZK7 A0A2Z5RDW3 A0A336LTU8 A0A3B0K067 A0A034VUQ7 A0A0K8VCG7 A0A0P5R8T6 A0A0P5XSB4 P41571 A0A182QEB5 A0A0K8UEE3 W8B8T2 A0A084WAQ4 A0A1Q3FEU9 A0A2J7QYS6 A0A2P8Y838 W8ATS3 A0A1D2NGI6 A0A1B6D356 B0WHG8 B7FNK0 A0A2J7QYR4 A0A182FUC7 A0A182J8K4 A0A182P1B9 W5JU82 A0A182WXU3 A0A182NDA4 X2JEF0 A0A182VKK6 A0A182KV52 H2KMF3 A0A0L7QRK3 A0A2M4AU73 A0A226F0P3 B3MQC4 P12646 A0A182Y6T9 A0A0P9A5S1 A0A182WFS6 Q0IEL8 A0A195BFM2 A0A2M4AVT8 P12646-2 A0A182I5W4 A0A1Y9GJ71 A0A182GD35 U4UL56 Q1WKS9 A0A1Y9GIQ5 A0A0U2VLP2 A0A182MVE5 A0A310S6H7 N6TQG0 A0A232F7X7 A0A1J1HGQ8 B3NVS1 Q1WKS8 B4JJG9 A0A158NQV6 A0A0Q5TGW9
PDB
5UKW
E-value=0,
Score=1718
Ontologies
PATHWAY
GO
PANTHER
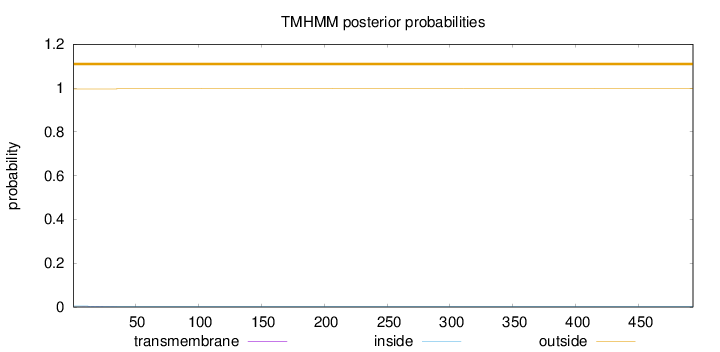
Topology
Length:
493
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03123
Exp number, first 60 AAs:
0.02941
Total prob of N-in:
0.00463
outside
1 - 493
Population Genetic Test Statistics
Pi
14.553743
Theta
37.726901
Tajima's D
-1.596032
CLR
0.646933
CSRT
0.0501474926253687
Interpretation
Uncertain
