Gene
KWMTBOMO01848
Pre Gene Modal
BGIBMGA006181
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_inactive_dipeptidyl_peptidase_10_[Bombyx_mori]
Location in the cell
Lysosomal Reliability : 1.342
Sequence
CDS
ATGGTCATAGGTTTTGTCATAGCCGGTATTGTGACTGCTATTTATTTATTAGGATATGTGGACGAATTACTATATTGGTCGGGTCGTCGTATGCGTTTGGACGAATTTCTGCGGGGCGATTTGACCGGAGAGCGTTTGCCGACCACCTGGGTGAGCTCTCACCAGCTCGTCTACCAGGCCGACGACGGAGGTCTACTGGCTCTGGACACCTTCAACAACACGCTCACTGTGCTCGTGACGAACCATACATTGAGACAACTAAACGTCCGATGGTACCAATGCTCACCGAATTTAAGATACGTACTGTTTCAACACAATATCAAAGAGGTTTATCGACGAACGTTTACAGCGTACTATACCGTTTATGACGTTACCAACGATCACCACATTCCACTGTTCGGAGAGGGACAGAGCAGCTGGGAGTGGCAACACGCGGCGTGGCTCGGTCCCGACGGGTCGCTGCTGCTAGCCGCTGACAACGAGGTGCTCGCAAGGCCCGGACCTCCCGCGCGGAGGGCACCCCTACTACGACTCACCAACGACTCCAGCCCTGGACGTATTTACAATGGAGTCTCAGACTGGTTGTATCAAGAGGAGGTGACGAAAACTTCATCCGCGACATGGGGTTCGATAGACGGCACGCTGGTGATGTACGTGCAGTACGACGACAGCAGCGTGTCCGAGCTCCGGTTCCCGCGCATCGCGGAGGGCATCGGCGGCGCCGGAGCCTCGCGGACCGGCTTCCTGCTGCCCACGCTCAACAACACGCACCACACCGTTTTTCCAGATCATGTTACTATCAGATATCCCACGCCTGGCAGTTCAATACCAAAAGTCAAACTATGGATTGTAGCTGTTCTGAATACTACATCACCTCCTCGATGGGAAGTGAAACCTCCGAGTACATTAGATGGAATGGAATATTACTTCATATCAGCCCAGTGGGTGGGCAAAGAGAATTCTCATGTAGGTGTAGTGTGGATGAACCGTGCACAGAATCTCACTGTTTATAGCAGTTGTTACGCTCCGAATTGGACATGTACAGAGTCACATTCAGAGAAAGCGACAGATGAGCCTTGGTTAGAAGTCCATCAACAGCCAGTATATTCAGAGGACGGTAGCGCTTTCCTCCTACTAGCTGCAGTTCAAGAGGGTGGAGGACAGTACTATACTCACATCAAGCACGTTGATGTCATACGACAACGTATAGCCGTGCTGTCGCATGGGAAAGTAGAAGTCGCCGAAATCCTGGCGTGGGACCAGGAAAATAATTTAGTTTATTATTTAGGCAGTGCAGATAGACCTGGTCAGAGACAGGTCTATGTGGTCCGAGACCCTAATTATAGCGGTGGAAGTAACTCGGTGAAAGCTCGAGCCGAAAGGGAAGAGCCGCGTTGCCTTACCTGTGAGCTGGCGGTGTGGCCCGCGCGGTTGCATTACGCCAACTGTTCATTCTGGCGCGCCATTTTCTCCCCGGCTAAGCCGAGAGTGGGGATAACGCATTACGTGCTGGAGTGCAGAGGTCCTGGACCACCATTAGCGGGGTTACATGATGCTAGGACACATAAGCTAGAAAGAATTTTATATGACACGAGGCCTTATAGATCAGTTCGATTGCGCGAGTTAGCGTTGCCTACTCGCCGGTCGTTCGAAGTACAGCTGGGCACGGGGTCCAAAGCTCGGGTCCAGCTGCTCCTGCCGCCTTCGTGGCGAGAGGAACTGCGTGATGCTGCCTTTCCGGTGCTTGTTCATGTCGACGGCAGACCGGGTAGCCAGCAAGTGACTGAGGAGTTTGTAGTGGATTGGGGCTCATACATGTCGTCTCGCAACGACGTGGTATACGTAAAGCTGGACGTGGCGGGTGCGCGCGGTCTACCCAGGGCCCTGCTGCGGGGTCGCCTCGGGGGAGTTGAAGTAGCTGACCAATTGGCTGTTATTAGATATTTGTTAGAAACGTTCAAGTTTTTAGACGTAACTAGAGTTGCTGTGTGGGGTTGGGGCTATGGTGGGTACGTGACGGCCATGTTGCTCGGCTCTCAGCAGACCACGCTGAAGTGCGGTATTGCTGTTTCGCCGATCACTGATTGGCTTTATTATAATGCAGCATTCACGGAGCGTATTTTGGGCAAACCGTCCGTGAACTACAAAGGCTACGTAGAGGCGGACGCATCTCAGCGTGCGCACCACGTGCCCGCGCATGCGCTGTACCTCATACACGGCATGGCTGATATGAGCGCTCCCTACCCGCACTCCCTGCAATTAGCCAGGGCGCTCACTGACGCCGGCGTGCTCTTTAGATATCAGGCCTACGCAGACGAAGGGCACGACTTAAAAGGCGTCATCGAGCACGTGTACCGTTCAATGGAAGACTTCCTTCGCGAGTGCCTCTCCCTCGACCCAGAAGACACCAAGTTGCCTCCTCCCGACAGATAG
Protein
MVIGFVIAGIVTAIYLLGYVDELLYWSGRRMRLDEFLRGDLTGERLPTTWVSSHQLVYQADDGGLLALDTFNNTLTVLVTNHTLRQLNVRWYQCSPNLRYVLFQHNIKEVYRRTFTAYYTVYDVTNDHHIPLFGEGQSSWEWQHAAWLGPDGSLLLAADNEVLARPGPPARRAPLLRLTNDSSPGRIYNGVSDWLYQEEVTKTSSATWGSIDGTLVMYVQYDDSSVSELRFPRIAEGIGGAGASRTGFLLPTLNNTHHTVFPDHVTIRYPTPGSSIPKVKLWIVAVLNTTSPPRWEVKPPSTLDGMEYYFISAQWVGKENSHVGVVWMNRAQNLTVYSSCYAPNWTCTESHSEKATDEPWLEVHQQPVYSEDGSAFLLLAAVQEGGGQYYTHIKHVDVIRQRIAVLSHGKVEVAEILAWDQENNLVYYLGSADRPGQRQVYVVRDPNYSGGSNSVKARAEREEPRCLTCELAVWPARLHYANCSFWRAIFSPAKPRVGITHYVLECRGPGPPLAGLHDARTHKLERILYDTRPYRSVRLRELALPTRRSFEVQLGTGSKARVQLLLPPSWREELRDAAFPVLVHVDGRPGSQQVTEEFVVDWGSYMSSRNDVVYVKLDVAGARGLPRALLRGRLGGVEVADQLAVIRYLLETFKFLDVTRVAVWGWGYGGYVTAMLLGSQQTTLKCGIAVSPITDWLYYNAAFTERILGKPSVNYKGYVEADASQRAHHVPAHALYLIHGMADMSAPYPHSLQLARALTDAGVLFRYQAYADEGHDLKGVIEHVYRSMEDFLRECLSLDPEDTKLPPPDR
Summary
Similarity
Belongs to the peptidase S9B family.
Uniprot
A0A194PLR4
A0A2A4JCJ7
A0A2H1WXA6
H9J9I8
A0A212F609
A0A0L7L3S5
+ More
A0A067RJJ1 A0A1B0FUX5 A0A182RUW6 A0A182K6V2 A0A182PWH1 A0A182X0Y0 A7UT73 A0A182QYS7 A0A084WKY4 A0A182WMA2 A0A0L7QVN2 W5JV04 A0A310SSB3 A0A088A1D1 A0A0A1X8E9 A0A2A3ENL0 A0A1Q3FUR8 A0A1B0ANZ0 A0A1I8MZZ6 A0A1A9XVU8 W8B1Z2 A0A1A9V2A1 A0A3L8DQQ5 A0A182YJ25 A0A1I8PJR0 A0A182HPE7 A0A0C9RBH7 A0A1A9W5C8 A0A1Y1M488 A0A0R3NX07 B4LR00 A0A1W4WYK1 A0A336LT98 A0A1W4WMI7 A0A3B0JGU8 A0A1B0G2A1 A0A154PM65 A0A0M4EFG6 E2AF07 A0A0Q9XDK8 A0A1B0A4S8 A0A158N9K8 A0A195BD39 A0A1J1HT75 A0A0P8XRT4 A0A0T6B6G7 A0A182J5A1 A0A0R1E7Q8 A0A182M8G9 B4JPQ3 A0A195C6V2 F4WLZ1 A0A195FU49 A0A0C4DHM8 D6WJ59 A0A182NLV6 A0A1B6E9X0 A0A1B6CFX6 A0A026WCG2 E0VF98 U4TY50 N6TPJ4 A0A023EYJ8 A0A195DCP6 A0A182FEN6 A0A1S4EL01 A0A3Q0J9N8 T1HY29 E2B8W0 A0A0Q5WQK7 A0A1B6DBA4 A0A0M9A083 A0A0P9AHM3 A0A0A9Y624 A0A0A9Y8N5 E9J377 A0A232EY97 A0A1B6LT15
A0A067RJJ1 A0A1B0FUX5 A0A182RUW6 A0A182K6V2 A0A182PWH1 A0A182X0Y0 A7UT73 A0A182QYS7 A0A084WKY4 A0A182WMA2 A0A0L7QVN2 W5JV04 A0A310SSB3 A0A088A1D1 A0A0A1X8E9 A0A2A3ENL0 A0A1Q3FUR8 A0A1B0ANZ0 A0A1I8MZZ6 A0A1A9XVU8 W8B1Z2 A0A1A9V2A1 A0A3L8DQQ5 A0A182YJ25 A0A1I8PJR0 A0A182HPE7 A0A0C9RBH7 A0A1A9W5C8 A0A1Y1M488 A0A0R3NX07 B4LR00 A0A1W4WYK1 A0A336LT98 A0A1W4WMI7 A0A3B0JGU8 A0A1B0G2A1 A0A154PM65 A0A0M4EFG6 E2AF07 A0A0Q9XDK8 A0A1B0A4S8 A0A158N9K8 A0A195BD39 A0A1J1HT75 A0A0P8XRT4 A0A0T6B6G7 A0A182J5A1 A0A0R1E7Q8 A0A182M8G9 B4JPQ3 A0A195C6V2 F4WLZ1 A0A195FU49 A0A0C4DHM8 D6WJ59 A0A182NLV6 A0A1B6E9X0 A0A1B6CFX6 A0A026WCG2 E0VF98 U4TY50 N6TPJ4 A0A023EYJ8 A0A195DCP6 A0A182FEN6 A0A1S4EL01 A0A3Q0J9N8 T1HY29 E2B8W0 A0A0Q5WQK7 A0A1B6DBA4 A0A0M9A083 A0A0P9AHM3 A0A0A9Y624 A0A0A9Y8N5 E9J377 A0A232EY97 A0A1B6LT15
Pubmed
26354079
19121390
22118469
26227816
24845553
12364791
+ More
14747013 17210077 24438588 20920257 23761445 25830018 25315136 24495485 30249741 25244985 28004739 15632085 17994087 23185243 18057021 20798317 21347285 17550304 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18362917 19820115 24508170 20566863 23537049 25474469 25401762 21282665 28648823
14747013 17210077 24438588 20920257 23761445 25830018 25315136 24495485 30249741 25244985 28004739 15632085 17994087 23185243 18057021 20798317 21347285 17550304 21719571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 18362917 19820115 24508170 20566863 23537049 25474469 25401762 21282665 28648823
EMBL
KQ459601
KPI93933.1
NWSH01001958
PCG69569.1
ODYU01011700
SOQ57606.1
+ More
BABH01004897 BABH01004898 AGBW02010086 OWR49175.1 JTDY01003123 KOB70112.1 KK852432 KDR23987.1 AJWK01006360 AAAB01008900 EDO64056.1 AXCN02000946 ATLV01024157 KE525350 KFB50878.1 KQ414725 KOC62683.1 ADMH02000413 ETN66609.1 KQ760945 OAD58605.1 GBXI01006915 JAD07377.1 KZ288215 PBC32786.1 GFDL01003694 JAV31351.1 JXJN01001081 GAMC01015502 JAB91053.1 QOIP01000005 RLU22775.1 APCN01002984 GBYB01005660 JAG75427.1 GEZM01043444 JAV79115.1 CH379059 KRT03771.1 KRT03772.1 KRT03773.1 CH940649 EDW64539.2 KRF81691.1 KRF81692.1 KRF81693.1 KRF81694.1 UFQT01000101 SSX19963.1 OUUW01000006 SPP81614.1 CCAG010004343 CCAG010004344 KQ434978 KZC12975.1 CP012523 ALC39180.1 GL438984 EFN67962.1 CH933807 KRG03831.1 KRG03832.1 KRG03833.1 KRG03834.1 KRG03835.1 ADTU01001396 ADTU01001397 ADTU01001398 KQ976519 KYM82105.1 CVRI01000020 CRK90580.1 CH902643 KPU77331.1 LJIG01009559 KRT82837.1 CH891640 KRK05065.1 KRK05067.1 AXCM01012850 CH916372 EDV98883.1 KQ978220 KYM96355.1 GL888217 EGI64635.1 KQ981264 KYN43951.1 AE013599 EAA46098.4 KQ971343 EFA04428.2 GEDC01002591 JAS34707.1 GEDC01024999 JAS12299.1 KK107293 EZA53371.1 DS235108 EEB12054.1 KB631820 ERL86544.1 APGK01059511 APGK01059512 APGK01059513 KB741294 ENN70191.1 GBBI01004520 JAC14192.1 KQ980989 KYN10653.1 ACPB03007950 ACPB03007951 GL446390 EFN87859.1 CH954177 KQS71080.1 GEDC01014423 JAS22875.1 KQ435803 KOX73169.1 KPU77333.1 GBHO01015082 JAG28522.1 GBHO01015085 JAG28519.1 GL768058 EFZ12728.1 NNAY01001609 OXU23436.1 GEBQ01013147 JAT26830.1
BABH01004897 BABH01004898 AGBW02010086 OWR49175.1 JTDY01003123 KOB70112.1 KK852432 KDR23987.1 AJWK01006360 AAAB01008900 EDO64056.1 AXCN02000946 ATLV01024157 KE525350 KFB50878.1 KQ414725 KOC62683.1 ADMH02000413 ETN66609.1 KQ760945 OAD58605.1 GBXI01006915 JAD07377.1 KZ288215 PBC32786.1 GFDL01003694 JAV31351.1 JXJN01001081 GAMC01015502 JAB91053.1 QOIP01000005 RLU22775.1 APCN01002984 GBYB01005660 JAG75427.1 GEZM01043444 JAV79115.1 CH379059 KRT03771.1 KRT03772.1 KRT03773.1 CH940649 EDW64539.2 KRF81691.1 KRF81692.1 KRF81693.1 KRF81694.1 UFQT01000101 SSX19963.1 OUUW01000006 SPP81614.1 CCAG010004343 CCAG010004344 KQ434978 KZC12975.1 CP012523 ALC39180.1 GL438984 EFN67962.1 CH933807 KRG03831.1 KRG03832.1 KRG03833.1 KRG03834.1 KRG03835.1 ADTU01001396 ADTU01001397 ADTU01001398 KQ976519 KYM82105.1 CVRI01000020 CRK90580.1 CH902643 KPU77331.1 LJIG01009559 KRT82837.1 CH891640 KRK05065.1 KRK05067.1 AXCM01012850 CH916372 EDV98883.1 KQ978220 KYM96355.1 GL888217 EGI64635.1 KQ981264 KYN43951.1 AE013599 EAA46098.4 KQ971343 EFA04428.2 GEDC01002591 JAS34707.1 GEDC01024999 JAS12299.1 KK107293 EZA53371.1 DS235108 EEB12054.1 KB631820 ERL86544.1 APGK01059511 APGK01059512 APGK01059513 KB741294 ENN70191.1 GBBI01004520 JAC14192.1 KQ980989 KYN10653.1 ACPB03007950 ACPB03007951 GL446390 EFN87859.1 CH954177 KQS71080.1 GEDC01014423 JAS22875.1 KQ435803 KOX73169.1 KPU77333.1 GBHO01015082 JAG28522.1 GBHO01015085 JAG28519.1 GL768058 EFZ12728.1 NNAY01001609 OXU23436.1 GEBQ01013147 JAT26830.1
Proteomes
UP000053268
UP000218220
UP000005204
UP000007151
UP000037510
UP000027135
+ More
UP000092461 UP000075900 UP000075881 UP000075885 UP000076407 UP000007062 UP000075886 UP000030765 UP000075920 UP000053825 UP000000673 UP000005203 UP000242457 UP000092460 UP000095301 UP000092443 UP000078200 UP000279307 UP000076408 UP000095300 UP000075840 UP000091820 UP000001819 UP000008792 UP000192223 UP000268350 UP000092444 UP000076502 UP000092553 UP000000311 UP000009192 UP000092445 UP000005205 UP000078540 UP000183832 UP000007801 UP000075880 UP000002282 UP000075883 UP000001070 UP000078542 UP000007755 UP000078541 UP000000803 UP000007266 UP000075884 UP000053097 UP000009046 UP000030742 UP000019118 UP000078492 UP000069272 UP000079169 UP000015103 UP000008237 UP000008711 UP000053105 UP000215335
UP000092461 UP000075900 UP000075881 UP000075885 UP000076407 UP000007062 UP000075886 UP000030765 UP000075920 UP000053825 UP000000673 UP000005203 UP000242457 UP000092460 UP000095301 UP000092443 UP000078200 UP000279307 UP000076408 UP000095300 UP000075840 UP000091820 UP000001819 UP000008792 UP000192223 UP000268350 UP000092444 UP000076502 UP000092553 UP000000311 UP000009192 UP000092445 UP000005205 UP000078540 UP000183832 UP000007801 UP000075880 UP000002282 UP000075883 UP000001070 UP000078542 UP000007755 UP000078541 UP000000803 UP000007266 UP000075884 UP000053097 UP000009046 UP000030742 UP000019118 UP000078492 UP000069272 UP000079169 UP000015103 UP000008237 UP000008711 UP000053105 UP000215335
PRIDE
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A194PLR4
A0A2A4JCJ7
A0A2H1WXA6
H9J9I8
A0A212F609
A0A0L7L3S5
+ More
A0A067RJJ1 A0A1B0FUX5 A0A182RUW6 A0A182K6V2 A0A182PWH1 A0A182X0Y0 A7UT73 A0A182QYS7 A0A084WKY4 A0A182WMA2 A0A0L7QVN2 W5JV04 A0A310SSB3 A0A088A1D1 A0A0A1X8E9 A0A2A3ENL0 A0A1Q3FUR8 A0A1B0ANZ0 A0A1I8MZZ6 A0A1A9XVU8 W8B1Z2 A0A1A9V2A1 A0A3L8DQQ5 A0A182YJ25 A0A1I8PJR0 A0A182HPE7 A0A0C9RBH7 A0A1A9W5C8 A0A1Y1M488 A0A0R3NX07 B4LR00 A0A1W4WYK1 A0A336LT98 A0A1W4WMI7 A0A3B0JGU8 A0A1B0G2A1 A0A154PM65 A0A0M4EFG6 E2AF07 A0A0Q9XDK8 A0A1B0A4S8 A0A158N9K8 A0A195BD39 A0A1J1HT75 A0A0P8XRT4 A0A0T6B6G7 A0A182J5A1 A0A0R1E7Q8 A0A182M8G9 B4JPQ3 A0A195C6V2 F4WLZ1 A0A195FU49 A0A0C4DHM8 D6WJ59 A0A182NLV6 A0A1B6E9X0 A0A1B6CFX6 A0A026WCG2 E0VF98 U4TY50 N6TPJ4 A0A023EYJ8 A0A195DCP6 A0A182FEN6 A0A1S4EL01 A0A3Q0J9N8 T1HY29 E2B8W0 A0A0Q5WQK7 A0A1B6DBA4 A0A0M9A083 A0A0P9AHM3 A0A0A9Y624 A0A0A9Y8N5 E9J377 A0A232EY97 A0A1B6LT15
A0A067RJJ1 A0A1B0FUX5 A0A182RUW6 A0A182K6V2 A0A182PWH1 A0A182X0Y0 A7UT73 A0A182QYS7 A0A084WKY4 A0A182WMA2 A0A0L7QVN2 W5JV04 A0A310SSB3 A0A088A1D1 A0A0A1X8E9 A0A2A3ENL0 A0A1Q3FUR8 A0A1B0ANZ0 A0A1I8MZZ6 A0A1A9XVU8 W8B1Z2 A0A1A9V2A1 A0A3L8DQQ5 A0A182YJ25 A0A1I8PJR0 A0A182HPE7 A0A0C9RBH7 A0A1A9W5C8 A0A1Y1M488 A0A0R3NX07 B4LR00 A0A1W4WYK1 A0A336LT98 A0A1W4WMI7 A0A3B0JGU8 A0A1B0G2A1 A0A154PM65 A0A0M4EFG6 E2AF07 A0A0Q9XDK8 A0A1B0A4S8 A0A158N9K8 A0A195BD39 A0A1J1HT75 A0A0P8XRT4 A0A0T6B6G7 A0A182J5A1 A0A0R1E7Q8 A0A182M8G9 B4JPQ3 A0A195C6V2 F4WLZ1 A0A195FU49 A0A0C4DHM8 D6WJ59 A0A182NLV6 A0A1B6E9X0 A0A1B6CFX6 A0A026WCG2 E0VF98 U4TY50 N6TPJ4 A0A023EYJ8 A0A195DCP6 A0A182FEN6 A0A1S4EL01 A0A3Q0J9N8 T1HY29 E2B8W0 A0A0Q5WQK7 A0A1B6DBA4 A0A0M9A083 A0A0P9AHM3 A0A0A9Y624 A0A0A9Y8N5 E9J377 A0A232EY97 A0A1B6LT15
PDB
1XFD
E-value=1.20183e-76,
Score=732
Ontologies
GO
Topology
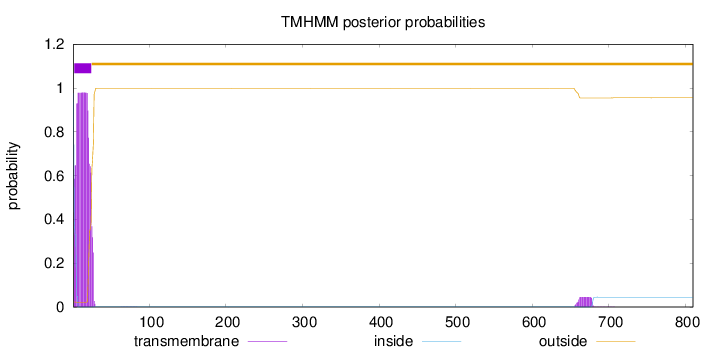
Length:
810
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.9523599999999
Exp number, first 60 AAs:
21.01082
Total prob of N-in:
0.97743
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 24
outside
25 - 810
Population Genetic Test Statistics
Pi
251.999243
Theta
186.349041
Tajima's D
1.422271
CLR
0.237559
CSRT
0.76271186440678
Interpretation
Uncertain
