Gene
KWMTBOMO01844 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA006180
Annotation
dipeptidyl-peptidase_[Danaus_plexippus]
Full name
Venom dipeptidyl peptidase 4
Alternative Name
Venom dipeptidyl peptidase IV
Allergen C
Allergen C
Location in the cell
PlasmaMembrane Reliability : 2.862
Sequence
CDS
ATGGAGCAAGCTGTCTTTCTGCTCATGGGTTTGATATGCCACACGCAATCGTCGGCATATATTCAGGGGCAAAAAATATTTTCATTAGAAGAAATAGTACCTCTGATGGCCGAGTTTTATCCTGAAAGAATGTCCGTGCAATGGATATCAGATACAACGTATATATACAAAGATCCTGGTGGGATAAAGAAATACGACGCCTCCACGGATAAAGTCGTCACGATTCTCAATGAAATCGAATTGTATAATTTGAGCAATTATACGCTGTCTACGTTTTCTGAAGACGGAAAATACGTGCTGCTAACGAAAAATAGAAGAAAGATATACAGATATTCATTCCTTGCGGAGTATTCCGTGCTAGATATTGAAAAGAGATTAATCTACAAGCTAAGCGATTATCCGCTACAAGTCGTGGTTTGGGGTATTGGGAAATCTGTAGCTTACGTTCAAGATAACAATGTATACTACGTACCTGACATTACCCAAGCAGATTATGTTGGTGTCCTCACCACAGGAGGAGTGCCCGGCGAAGTATATTTCGGTGCTACTGACTGGATTTACGAAGAGGAGATATTTAATGCAGCAGAAGCATTATGGTTTTCTCCGGGAGGTACTTACCTCGCTGTTGCTTCCTTCAATGATACTAACGTTGAATCGGCCGTATTCCCTTATTACGGAGATCCTGGCAACATAAACAACCAGTACCCACAAATGGTGCAGTTTAAATATCCTAAGGTAGGTCGGACAAATCCGAGAGTGGGTTTACGCGTCTACAAACTGGACGAGCCCAATAGTGAGCCTTGGAGTATACCTGCACCGGTCGATGTGATTGGACTCGACAACATCCTCGGTCGTGTAAACTGGGCTTCAGACCAAAATCTAATTGTTCTCTGGCTGAACAGACGACAAAGTATTAGTATCCTGATCAATTGTGACTTGCAAAAAGACAAATGTAGCATAGTGAAAGAACACACGGAACCAAACGGTTGGATCGACATTAACGACCCAATTTTCGACAAAACTGGAACTAAAATGGTGGAGATACAGCCTTTGTATCATGCTGAACGTCGTTTCCCGCACGCCGCAAGGGTCGATTTCAATACTTTAACAACTGAAGATCTGAGTCCCGGTAACTCTACGGTTTTGGAAATTGTTGGCTGGAATCAAGATACCAATACAGTGTATTACATAGTAGCTCCTGGAAATTTACCATGGCTAAGGCAAATATGGGCAACTTCCGGCGGTGTGGTTAGATGTATCTCGTGCAAAGAACCGACTTGTAGGCACGTGTCGGCCATGTTTTCACCGGGAGCCAACTACGCCATAGTAACGTGTAGCGCCTGCAACATCCCTCCTAAAATATACCTGTATGATGCCAAGAAAGAGACATTCAAATTGATAAAAGACAACAAAAAACTCATTGATACCCTACAAATGTACCAACTACCTCTTACCCATTTCATCGCAATGTCAATAGGAAACGAAGTGAATGCAAACGTGAAACTCATTCTTCCAGCTGGGATAAAGCAAGGAGAACAGTATGCAATGGTGGTTAGAGTGTACGCTGGTCCCGGGACAACCCGAGTTCGCGATAACTACGATCTAGAATATTACAACTCGTACCTGGCTACAAACAGGAGTATAATAGTGGCCTCGATCGACGTACGAGGTTCAGGAGTGATGGGCGTGGAAGCCATGCATGCAGTCAACAGGGCTTTGGGAACTGTCGAAATATCCGATACTTTGGATACAATCAAGAGCTTAACTAGTTTGTATTCTTTCATCGACCCTGACCGGATAGGAGTGTGGGGCTGGAGTTACGGTGGCTATGCCACCACAATGATGTTGATTCAGGATCAGCGGAACATTGTGAAATGTGGGGCCGCTGTCGCCCCCGTTACCTCATGGCTCTATTACGATTCAATATACACGGAACGTTATATGGATACGCCTCAAGCGAACCCTTTGGGCTACGAACGATCTGACCTAACTAATCTAGCTGAAGGATTGCGAGGACGCAAATATTTGCTGGTTCACGGGAGCGGTGACGACAATGTGCATTACCAGCACAGCATGCAACTCGCCAAGCGATTGCAGCATAGCGATATTGCCTTCGAGCAAATGAGCTATACGGACGAGAATCATTCCTTGATGGGAGTGAGTCGACATTTCTACCACACGTTGGATCGCTTCTGGTCAGAGTGCTTCAAATTATAA
Protein
MEQAVFLLMGLICHTQSSAYIQGQKIFSLEEIVPLMAEFYPERMSVQWISDTTYIYKDPGGIKKYDASTDKVVTILNEIELYNLSNYTLSTFSEDGKYVLLTKNRRKIYRYSFLAEYSVLDIEKRLIYKLSDYPLQVVVWGIGKSVAYVQDNNVYYVPDITQADYVGVLTTGGVPGEVYFGATDWIYEEEIFNAAEALWFSPGGTYLAVASFNDTNVESAVFPYYGDPGNINNQYPQMVQFKYPKVGRTNPRVGLRVYKLDEPNSEPWSIPAPVDVIGLDNILGRVNWASDQNLIVLWLNRRQSISILINCDLQKDKCSIVKEHTEPNGWIDINDPIFDKTGTKMVEIQPLYHAERRFPHAARVDFNTLTTEDLSPGNSTVLEIVGWNQDTNTVYYIVAPGNLPWLRQIWATSGGVVRCISCKEPTCRHVSAMFSPGANYAIVTCSACNIPPKIYLYDAKKETFKLIKDNKKLIDTLQMYQLPLTHFIAMSIGNEVNANVKLILPAGIKQGEQYAMVVRVYAGPGTTRVRDNYDLEYYNSYLATNRSIIVASIDVRGSGVMGVEAMHAVNRALGTVEISDTLDTIKSLTSLYSFIDPDRIGVWGWSYGGYATTMMLIQDQRNIVKCGAAVAPVTSWLYYDSIYTERYMDTPQANPLGYERSDLTNLAEGLRGRKYLLVHGSGDDNVHYQHSMQLAKRLQHSDIAFEQMSYTDENHSLMGVSRHFYHTLDRFWSECFKL
Summary
Description
Venom dipeptidyl-peptidase which removes N-terminal dipeptides sequentially from polypeptides having unsubstituted N-termini provided that the penultimate residue is proline. May process venom proteins into their active forms and/or modulate the chemotactic activity of immune cells after the insect sting.
Venom dipeptidyl-peptidase which removes N-terminal dipeptides sequentially from polypeptides having unsubstituted N-termini provided that the penultimate residue is proline. May process promelittin into its active form and/or modulate the chemotactic activity of immune cells after the insect sting.
Venom dipeptidyl-peptidase which removes N-terminal dipeptides sequentially from polypeptides having unsubstituted N-termini provided that the penultimate residue is proline. May process promelittin into its active form and/or modulate the chemotactic activity of immune cells after the insect sting.
Catalytic Activity
Release of an N-terminal dipeptide, Xaa-Yaa-|-Zaa-, from a polypeptide, preferentially when Yaa is Pro, provided Zaa is neither Pro nor hydroxyproline.
Similarity
Belongs to the peptidase S9B family.
Belongs to the peptidase S9B family. DPPIV subfamily.
Belongs to the peptidase S9B family. DPPIV subfamily.
Keywords
Allergen
Aminopeptidase
Direct protein sequencing
Disulfide bond
Glycoprotein
Hydrolase
Protease
Secreted
Signal
Complete proteome
Reference proteome
Feature
chain Venom dipeptidyl peptidase 4
Uniprot
H9J9I7
A0A437BSW5
A0A2A4JD71
A0A2H1W553
A0A194PS22
A0A212F613
+ More
A0A0L7LI97 A0A194R3X6 A0A3L8DR73 E2AF09 A0A026WEB2 A0A2J7Q7T3 Q16EL4 F4WLY9 A0A1Q3FKC6 Q1DH03 B0WE34 A0A158N9M0 B1A4F7 A4UA14 A0A182JFB8 A0A023EV00 A0A348G603 V9PBD6 A0A182HC95 A0A084W3B1 A0A0P6JXV0 A0A1B6DL67 A0A212F622 A0A1B6EG93 A0A0P6A850 A0A182PPN8 A0A195BBI8 A0A164UG38 A0A0N8BLW1 A0A1B6FT22 A0A1B6HM76 A0A1B6ICV7 A0A182YMC6 A0A0P6E1N2 A0A0P5YLG7 E9FZG6 A0A0P4W4J7 A0A0V0G4N0 A0A023EXQ5 E2B8W2 A0A069DX38 A0A195FVS0 A0A195DE08 A0A023EZT7 A0A182NFA4 A0A1L8DPB2 A0A1S4H707 Q7PSF9 A0A182URD4 A0A182UKA9 A0A1B6DL51 A0A1L8DPL6 A0A146KL11 A0A310SPV5 A0A0A9XS95 A0A195C860 A0A182X7R6 A0A182LP15 H9J9I6 W5JGF1 A0A023EVT1 A0A336M465 A0A232ETT6 A0A182HN75 A0A0K8SCK7 D6WJ57 A0A0P5M9I8 A0A088A1D0 A0A023EVG2 B2D0J4 K7JAQ2 A0A1B6GG80 U5EWK1 A0A1Y1KXZ8 A0A1B0CY21 A0A2M4A0T9 A0A2M4A0V0 A0A2M4A0X2 A0A1W4U9C0 A0A0M4EGC5 A0A1L8EGB2 A0A1L8EFV7 Q16RL5 A0A2M4BE95 A0A182FBI4 A0A2M4BE23 A0A3R7LWT5 A0A182Y621 A0A0R1E3Q4 A0A1L8EG47 A0A0R3P7Z3 B3N577 A0A0J9RVJ9 A0A1L8EG80
A0A0L7LI97 A0A194R3X6 A0A3L8DR73 E2AF09 A0A026WEB2 A0A2J7Q7T3 Q16EL4 F4WLY9 A0A1Q3FKC6 Q1DH03 B0WE34 A0A158N9M0 B1A4F7 A4UA14 A0A182JFB8 A0A023EV00 A0A348G603 V9PBD6 A0A182HC95 A0A084W3B1 A0A0P6JXV0 A0A1B6DL67 A0A212F622 A0A1B6EG93 A0A0P6A850 A0A182PPN8 A0A195BBI8 A0A164UG38 A0A0N8BLW1 A0A1B6FT22 A0A1B6HM76 A0A1B6ICV7 A0A182YMC6 A0A0P6E1N2 A0A0P5YLG7 E9FZG6 A0A0P4W4J7 A0A0V0G4N0 A0A023EXQ5 E2B8W2 A0A069DX38 A0A195FVS0 A0A195DE08 A0A023EZT7 A0A182NFA4 A0A1L8DPB2 A0A1S4H707 Q7PSF9 A0A182URD4 A0A182UKA9 A0A1B6DL51 A0A1L8DPL6 A0A146KL11 A0A310SPV5 A0A0A9XS95 A0A195C860 A0A182X7R6 A0A182LP15 H9J9I6 W5JGF1 A0A023EVT1 A0A336M465 A0A232ETT6 A0A182HN75 A0A0K8SCK7 D6WJ57 A0A0P5M9I8 A0A088A1D0 A0A023EVG2 B2D0J4 K7JAQ2 A0A1B6GG80 U5EWK1 A0A1Y1KXZ8 A0A1B0CY21 A0A2M4A0T9 A0A2M4A0V0 A0A2M4A0X2 A0A1W4U9C0 A0A0M4EGC5 A0A1L8EGB2 A0A1L8EFV7 Q16RL5 A0A2M4BE95 A0A182FBI4 A0A2M4BE23 A0A3R7LWT5 A0A182Y621 A0A0R1E3Q4 A0A1L8EG47 A0A0R3P7Z3 B3N577 A0A0J9RVJ9 A0A1L8EG80
EC Number
3.4.14.5
Pubmed
19121390
26354079
22118469
26227816
30249741
20798317
+ More
24508170 17510324 21719571 21347285 20348419 17298553 24945155 26483478 24438588 25244985 21292972 27129103 26334808 25474469 12364791 26823975 25401762 20966253 20920257 23761445 28648823 18362917 19820115 17073008 20075255 28004739 17994087 17550304 15632085 22936249
24508170 17510324 21719571 21347285 20348419 17298553 24945155 26483478 24438588 25244985 21292972 27129103 26334808 25474469 12364791 26823975 25401762 20966253 20920257 23761445 28648823 18362917 19820115 17073008 20075255 28004739 17994087 17550304 15632085 22936249
EMBL
BABH01004877
RSAL01000014
RVE53279.1
NWSH01001848
PCG69921.1
ODYU01006093
+ More
SOQ47634.1 KQ459601 KPI93935.1 AGBW02010086 OWR49177.1 JTDY01001074 KOB74961.1 KQ460855 KPJ11945.1 QOIP01000005 RLU22782.1 GL438984 EFN67964.1 KK107293 EZA53369.1 NEVH01017440 PNF24641.1 CH478666 EAT32671.1 GL888217 EGI64633.1 GFDL01007083 JAV27962.1 CH899921 EAT32433.1 DS231904 EDS45210.1 ADTU01001408 EU420987 DQ661743 ABG57265.1 GAPW01000318 JAC13280.1 FX985542 BBF97876.1 KC282404 AGX25161.1 JXUM01126919 KQ567125 KXJ69612.1 ATLV01019900 KE525284 KFB44705.1 GDIQ01004025 JAN90712.1 GEDC01010864 JAS26434.1 OWR49178.1 GEDC01000404 JAS36894.1 GDIP01033893 JAM69822.1 KQ976528 KYM81921.1 LRGB01001581 KZS11328.1 GDIQ01164971 GDIQ01067404 JAK86754.1 GECZ01016434 JAS53335.1 GECU01031918 JAS75788.1 GECU01022932 JAS84774.1 GDIQ01070450 JAN24287.1 GDIP01056093 JAM47622.1 GL732528 EFX87272.1 GDKW01000335 JAI56260.1 GECL01003131 JAP02993.1 GAPW01000319 JAC13279.1 GL446390 EFN87861.1 GBGD01000523 JAC88366.1 KQ981264 KYN43954.1 KQ980989 KYN10654.1 GBBI01003949 JAC14763.1 GFDF01005805 JAV08279.1 AAAB01008834 EAA05700.5 GEDC01010885 GEDC01001959 JAS26413.1 JAS35339.1 GFDF01005804 JAV08280.1 GDHC01021358 JAP97270.1 KQ760343 OAD60840.1 GBHO01021085 GBHO01021084 GBHO01021083 GBHO01015182 JAG22519.1 JAG22520.1 JAG22521.1 JAG28422.1 KQ978220 KYM96356.1 BABH01004876 ADMH02001286 ETN63166.1 GAPW01000407 JAC13191.1 UFQS01000538 UFQT01000538 SSX04703.1 SSX25066.1 NNAY01002223 OXU21775.1 APCN01001877 GBRD01015321 JAG50505.1 KQ971343 EFA04427.1 GDIQ01182517 JAK69208.1 GAPW01000606 JAC12992.1 EU564832 AAZX01013515 GECZ01013355 GECZ01008349 JAS56414.1 JAS61420.1 GANO01000531 JAB59340.1 GEZM01070829 JAV66262.1 AJWK01035258 AJWK01035259 GGFK01001058 MBW34379.1 GGFK01001051 MBW34372.1 GGFK01001049 MBW34370.1 CP012523 ALC39730.1 GFDG01001186 JAV17613.1 GFDG01001189 JAV17610.1 CH477704 EAT37053.1 GGFJ01002221 MBW51362.1 GGFJ01002151 MBW51292.1 QCYY01002917 ROT66607.1 CM000159 KRK01895.1 GFDG01001183 JAV17616.1 CH379069 KRT07558.1 CH954177 EDV57907.1 CM002912 KMY99602.1 GFDG01001190 JAV17609.1
SOQ47634.1 KQ459601 KPI93935.1 AGBW02010086 OWR49177.1 JTDY01001074 KOB74961.1 KQ460855 KPJ11945.1 QOIP01000005 RLU22782.1 GL438984 EFN67964.1 KK107293 EZA53369.1 NEVH01017440 PNF24641.1 CH478666 EAT32671.1 GL888217 EGI64633.1 GFDL01007083 JAV27962.1 CH899921 EAT32433.1 DS231904 EDS45210.1 ADTU01001408 EU420987 DQ661743 ABG57265.1 GAPW01000318 JAC13280.1 FX985542 BBF97876.1 KC282404 AGX25161.1 JXUM01126919 KQ567125 KXJ69612.1 ATLV01019900 KE525284 KFB44705.1 GDIQ01004025 JAN90712.1 GEDC01010864 JAS26434.1 OWR49178.1 GEDC01000404 JAS36894.1 GDIP01033893 JAM69822.1 KQ976528 KYM81921.1 LRGB01001581 KZS11328.1 GDIQ01164971 GDIQ01067404 JAK86754.1 GECZ01016434 JAS53335.1 GECU01031918 JAS75788.1 GECU01022932 JAS84774.1 GDIQ01070450 JAN24287.1 GDIP01056093 JAM47622.1 GL732528 EFX87272.1 GDKW01000335 JAI56260.1 GECL01003131 JAP02993.1 GAPW01000319 JAC13279.1 GL446390 EFN87861.1 GBGD01000523 JAC88366.1 KQ981264 KYN43954.1 KQ980989 KYN10654.1 GBBI01003949 JAC14763.1 GFDF01005805 JAV08279.1 AAAB01008834 EAA05700.5 GEDC01010885 GEDC01001959 JAS26413.1 JAS35339.1 GFDF01005804 JAV08280.1 GDHC01021358 JAP97270.1 KQ760343 OAD60840.1 GBHO01021085 GBHO01021084 GBHO01021083 GBHO01015182 JAG22519.1 JAG22520.1 JAG22521.1 JAG28422.1 KQ978220 KYM96356.1 BABH01004876 ADMH02001286 ETN63166.1 GAPW01000407 JAC13191.1 UFQS01000538 UFQT01000538 SSX04703.1 SSX25066.1 NNAY01002223 OXU21775.1 APCN01001877 GBRD01015321 JAG50505.1 KQ971343 EFA04427.1 GDIQ01182517 JAK69208.1 GAPW01000606 JAC12992.1 EU564832 AAZX01013515 GECZ01013355 GECZ01008349 JAS56414.1 JAS61420.1 GANO01000531 JAB59340.1 GEZM01070829 JAV66262.1 AJWK01035258 AJWK01035259 GGFK01001058 MBW34379.1 GGFK01001051 MBW34372.1 GGFK01001049 MBW34370.1 CP012523 ALC39730.1 GFDG01001186 JAV17613.1 GFDG01001189 JAV17610.1 CH477704 EAT37053.1 GGFJ01002221 MBW51362.1 GGFJ01002151 MBW51292.1 QCYY01002917 ROT66607.1 CM000159 KRK01895.1 GFDG01001183 JAV17616.1 CH379069 KRT07558.1 CH954177 EDV57907.1 CM002912 KMY99602.1 GFDG01001190 JAV17609.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000037510
+ More
UP000053240 UP000279307 UP000000311 UP000053097 UP000235965 UP000008820 UP000007755 UP000002320 UP000005205 UP000075880 UP000069940 UP000249989 UP000030765 UP000075885 UP000078540 UP000076858 UP000076408 UP000000305 UP000008237 UP000078541 UP000078492 UP000075884 UP000007062 UP000075903 UP000075902 UP000078542 UP000076407 UP000075882 UP000000673 UP000215335 UP000075840 UP000007266 UP000005203 UP000002358 UP000092461 UP000192221 UP000092553 UP000069272 UP000283509 UP000002282 UP000001819 UP000008711
UP000053240 UP000279307 UP000000311 UP000053097 UP000235965 UP000008820 UP000007755 UP000002320 UP000005205 UP000075880 UP000069940 UP000249989 UP000030765 UP000075885 UP000078540 UP000076858 UP000076408 UP000000305 UP000008237 UP000078541 UP000078492 UP000075884 UP000007062 UP000075903 UP000075902 UP000078542 UP000076407 UP000075882 UP000000673 UP000215335 UP000075840 UP000007266 UP000005203 UP000002358 UP000092461 UP000192221 UP000092553 UP000069272 UP000283509 UP000002282 UP000001819 UP000008711
Interpro
Gene 3D
ProteinModelPortal
H9J9I7
A0A437BSW5
A0A2A4JD71
A0A2H1W553
A0A194PS22
A0A212F613
+ More
A0A0L7LI97 A0A194R3X6 A0A3L8DR73 E2AF09 A0A026WEB2 A0A2J7Q7T3 Q16EL4 F4WLY9 A0A1Q3FKC6 Q1DH03 B0WE34 A0A158N9M0 B1A4F7 A4UA14 A0A182JFB8 A0A023EV00 A0A348G603 V9PBD6 A0A182HC95 A0A084W3B1 A0A0P6JXV0 A0A1B6DL67 A0A212F622 A0A1B6EG93 A0A0P6A850 A0A182PPN8 A0A195BBI8 A0A164UG38 A0A0N8BLW1 A0A1B6FT22 A0A1B6HM76 A0A1B6ICV7 A0A182YMC6 A0A0P6E1N2 A0A0P5YLG7 E9FZG6 A0A0P4W4J7 A0A0V0G4N0 A0A023EXQ5 E2B8W2 A0A069DX38 A0A195FVS0 A0A195DE08 A0A023EZT7 A0A182NFA4 A0A1L8DPB2 A0A1S4H707 Q7PSF9 A0A182URD4 A0A182UKA9 A0A1B6DL51 A0A1L8DPL6 A0A146KL11 A0A310SPV5 A0A0A9XS95 A0A195C860 A0A182X7R6 A0A182LP15 H9J9I6 W5JGF1 A0A023EVT1 A0A336M465 A0A232ETT6 A0A182HN75 A0A0K8SCK7 D6WJ57 A0A0P5M9I8 A0A088A1D0 A0A023EVG2 B2D0J4 K7JAQ2 A0A1B6GG80 U5EWK1 A0A1Y1KXZ8 A0A1B0CY21 A0A2M4A0T9 A0A2M4A0V0 A0A2M4A0X2 A0A1W4U9C0 A0A0M4EGC5 A0A1L8EGB2 A0A1L8EFV7 Q16RL5 A0A2M4BE95 A0A182FBI4 A0A2M4BE23 A0A3R7LWT5 A0A182Y621 A0A0R1E3Q4 A0A1L8EG47 A0A0R3P7Z3 B3N577 A0A0J9RVJ9 A0A1L8EG80
A0A0L7LI97 A0A194R3X6 A0A3L8DR73 E2AF09 A0A026WEB2 A0A2J7Q7T3 Q16EL4 F4WLY9 A0A1Q3FKC6 Q1DH03 B0WE34 A0A158N9M0 B1A4F7 A4UA14 A0A182JFB8 A0A023EV00 A0A348G603 V9PBD6 A0A182HC95 A0A084W3B1 A0A0P6JXV0 A0A1B6DL67 A0A212F622 A0A1B6EG93 A0A0P6A850 A0A182PPN8 A0A195BBI8 A0A164UG38 A0A0N8BLW1 A0A1B6FT22 A0A1B6HM76 A0A1B6ICV7 A0A182YMC6 A0A0P6E1N2 A0A0P5YLG7 E9FZG6 A0A0P4W4J7 A0A0V0G4N0 A0A023EXQ5 E2B8W2 A0A069DX38 A0A195FVS0 A0A195DE08 A0A023EZT7 A0A182NFA4 A0A1L8DPB2 A0A1S4H707 Q7PSF9 A0A182URD4 A0A182UKA9 A0A1B6DL51 A0A1L8DPL6 A0A146KL11 A0A310SPV5 A0A0A9XS95 A0A195C860 A0A182X7R6 A0A182LP15 H9J9I6 W5JGF1 A0A023EVT1 A0A336M465 A0A232ETT6 A0A182HN75 A0A0K8SCK7 D6WJ57 A0A0P5M9I8 A0A088A1D0 A0A023EVG2 B2D0J4 K7JAQ2 A0A1B6GG80 U5EWK1 A0A1Y1KXZ8 A0A1B0CY21 A0A2M4A0T9 A0A2M4A0V0 A0A2M4A0X2 A0A1W4U9C0 A0A0M4EGC5 A0A1L8EGB2 A0A1L8EFV7 Q16RL5 A0A2M4BE95 A0A182FBI4 A0A2M4BE23 A0A3R7LWT5 A0A182Y621 A0A0R1E3Q4 A0A1L8EG47 A0A0R3P7Z3 B3N577 A0A0J9RVJ9 A0A1L8EG80
PDB
4KR0
E-value=1.74902e-102,
Score=954
Ontologies
KEGG
GO
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 19,
Likelihood: 0.975971
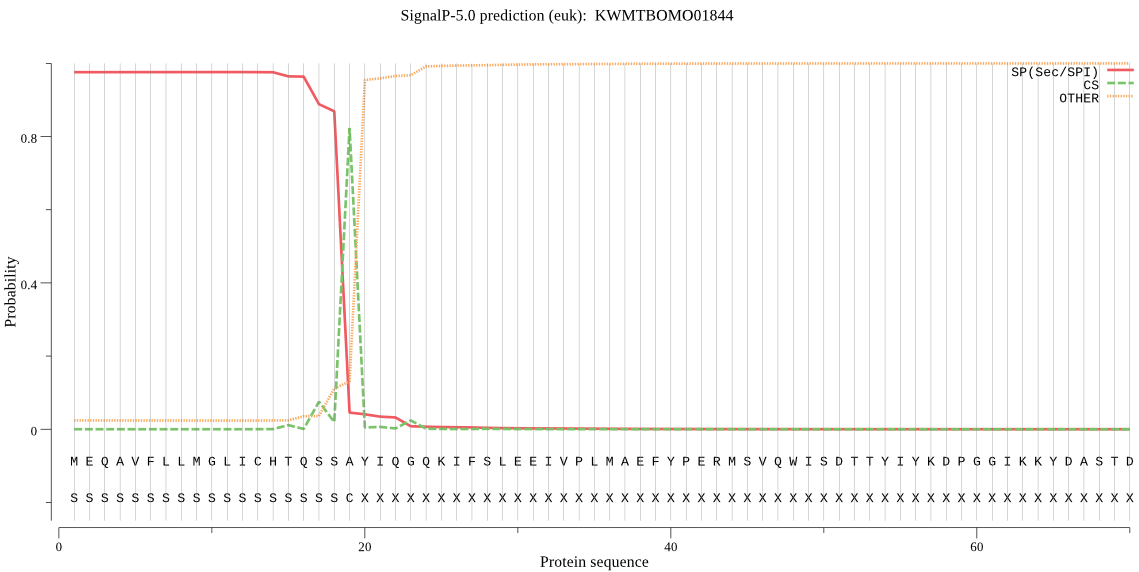
Length:
738
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10109
Exp number, first 60 AAs:
0.02591
Total prob of N-in:
0.00147
outside
1 - 738
Population Genetic Test Statistics
Pi
257.881386
Theta
181.963068
Tajima's D
1.150692
CLR
0.625463
CSRT
0.699915004249787
Interpretation
Uncertain
